Generic 4: SEIR - Vital dynamics (birth and death)¶
This tutorial assumes that you have read Generic 3: SIER - Incubation periods.
This tutorial explores the effect of birth and death on a simulation. In a closed population with no vital dynamics, an epidemic will eventually die out due to an insufficient number of susceptible individuals to sustain the disease. Infected individuals who are added later will not start another epidemic due to the life-long immunity of the existing population.
If there are vital dynamics, new births can provide more susceptible individuals to the population. In this case, when infected individuals are added later, another epidemic may start.
Demographics inputs¶
In the demographics input file, the node has 10,000 individuals. The birth rate and death rate are equal to 20 per 1000 individuals per year (0.0000548 per day).
The birth rate and initial population values are set in NodeAttributes using BirthRate and InitialPopulation, respectively. The death rate is set in IndividualAttributes using the MortalityDistribution. Both the male and female death rate is equal to 0.0000548 per day.
{
"Metadata": {
},
"Nodes": [{
"NodeID": 1,
"NodeAttributes": {
"InitialPopulation": 10000,
"BirthRate": 0.0000548
},
"IndividualAttributes": {
"MortalityDistribution": {
"NumDistributionAxes": 2,
"AxisNames": ["gender", "age"],
"AxisUnits": ["male=0,female=1", "years"],
"AxisScaleFactors": [1, 365],
"NumPopulationGroups": [2, 1],
"PopulationGroups": [
[0, 1],
[0]
],
"ResultUnits": "deaths per day",
"ResultScaleFactor": 1,
"ResultValues": [
[0.0000548],
[0.0000548]
]
}
}
}]
}
To view the complete demographics file, see generic_scenarios_demographics at <path_to_directory>RegressionScenariosInputFiles where <path_to_directory> is the location where EMOD source files were installed. For more information on demographics files, see Demographics parameters.
Key simulation parameters¶
This tutorial simulates the person-person disease transmission as an SEIR model in a hypothetical community over a longer period.
You can view the complete config.json in <path_to_directory>RegressionScenariosGeneric04_VitalDynamics directory.
Population dynamics parameters¶
The following parameters enable vital dynamics for birth and non-disease related deaths, and disable disease-related mortality. For more information, see Population dynamics parameters.
Set the following parameters:
- Set Enable_Vital_Dynamics to 1.
- Set Enable_Birth to 1.
- Set Death_Rate_Dependence to “NONDISEASE_MORTALITY_BY_AGE_AND_GENDER.”
- Set Enable_Disease_Mortality to 0.
SEIR model setup parameters¶
The incubation period, infection period, and immunity must be enabled for an SEIR model. Immunity waning must be disabled. For more information, see Incubation and Immunity parameters.
Set the following parameters:
- Set Base_Incubation_Period to 3.
- Set Enable_Immunity to 1.
- Set Enable_Immune_Decay to 0.
- Set Incubation_Period_Distribution to “EXPONENTIAL_DISTRIBUTION.”
Disease parameters¶
These parameters configure an SEIR-like disease model with person-to-person direct transmission. The basic reproductive number is 17.5.
The birth and death rates are calculated based on the base birth and mortality rates in the demographics file, as well as the multiplier in the configuration file. The birth and death (mortality) rates are increased by a factor of 10 using multipliers, x_Birth and x_Other_Mortality, to show the effect of the vital process. In this tutorial, the birth rate equals the mortality rate. For more information, see Infectivity and transmission and Scalars and multipliers parameters.
Set the following parameters:
- Set Base_Infectivity to 2.5.
- Set Base_Infectious_Period to 7.
- Set Infectious_Period_Distribution to “EXPONENTIAL_DURATION.”
- Set x_Birth to 10.
- Set x_Other_Mortality to 10.
Simulation duration parameters¶
The simulation runs a 10 year simulation with a 1-day time step. For more information, see Simulation setup parameters.
Set the following parameters:
- Set Simulation_Timestep to 1.
- Set Simulation_Duration to 3650.
Interventions¶
Prior immunity¶
This tutorial assumes that 80% of individuals are immune at the beginning of the simulation. This is simulated using a vaccination campaign with 80% coverage and 100% take at day 1.
{
"Campaign_Name": "Pre-outbreak Immunity",
"Event_Coordinator_Config": {
"Demographic_Coverage": 0.8,
"Intervention_Config": {
"Cost_To_Consumer": 10.0,
"Durability_Time_Profile": "BOXDURABILITY",
"Primary_Decay_Time_Constant": 3650,
"Reduced_Acquire": 1,
"Reduced_Transmit": 0,
"Vaccine_Take": 1,
"Vaccine_Type": "AcquisitionBlocking",
"class": "SimpleVaccine"
},
"Target_Demographic": "Everyone",
"class": "StandardInterventionDistributionEventCoordinator"
},
"Nodeset_Config": {
"class": "NodeSetAll"
},
"Start_Day": 1,
"class": "CampaignEvent"
}
Outbreaks¶
This simulation uses an OutbreakIndividual event for an infrequent infection seeding. The Timesteps_Between_Repetitions and Number_Repetitions parameters are set to reintroduce the outbreak once every two years for 10 years. With larger population sizes, the endemic can sustain itself. For more information, see OutbreakIndividual parameters.
{
"Campaign_Name": "Outbreak",
"Event_Coordinator_Config": {
"Demographic_Coverage": 0.001,
"Intervention_Config": {
"Antigen": 0,
"Event_Name": "Outbreak",
"Genome": 0,
"Outbreak_Source": "ImportCases",
"class": "OutbreakIndividual"
},
"Number_Repetitions": 10,
"Target_Demographic": "Everyone",
"Timesteps_Between_Repetitions": 730,
"class": "StandardInterventionDistributionEventCoordinator"
},
"Nodeset_Config": {
"class": "NodeSetAll"
},
"Start_Day": 5,
"class": "CampaignEvent"
}
You can view the complete campaign.json in <path_to_directory>RegressionScenariosGeneric04_VitalDynamics directory.
Run the simulation¶
Run the simulation and generate graphs of the simulation output. For more information, see Run simulations.
Note
Because the EMOD model is stochastic, your graphs may appear slightly different from those given below.
Simulation output graphs¶
The output shows the periodic outbreaks when an infection is reseeded because of the accumulation of new susceptibles due to the vital process (new births). Since periodic endemic behavior cannot be maintained due to stochastic fadeout in a small population, OutbreakIndividual was used to reintroduce the outbreak every two years for 10 years.
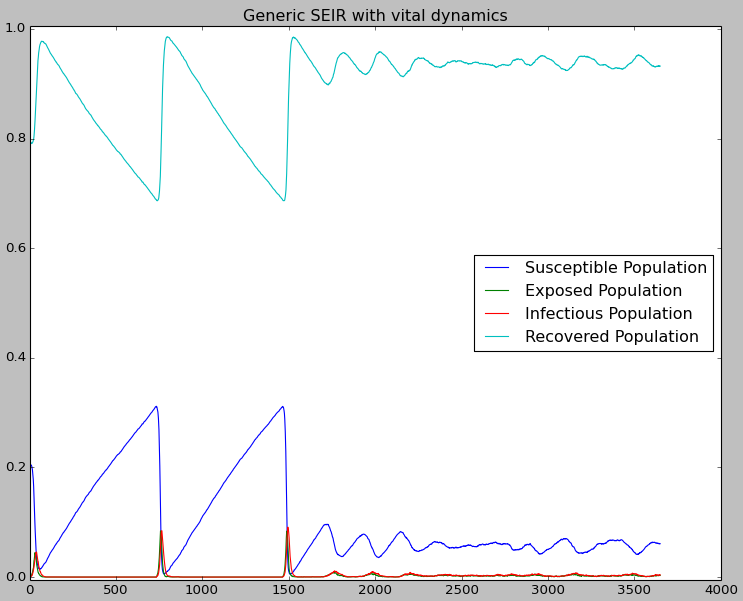
Figure 1: Periodic outbreaks on reintroduction
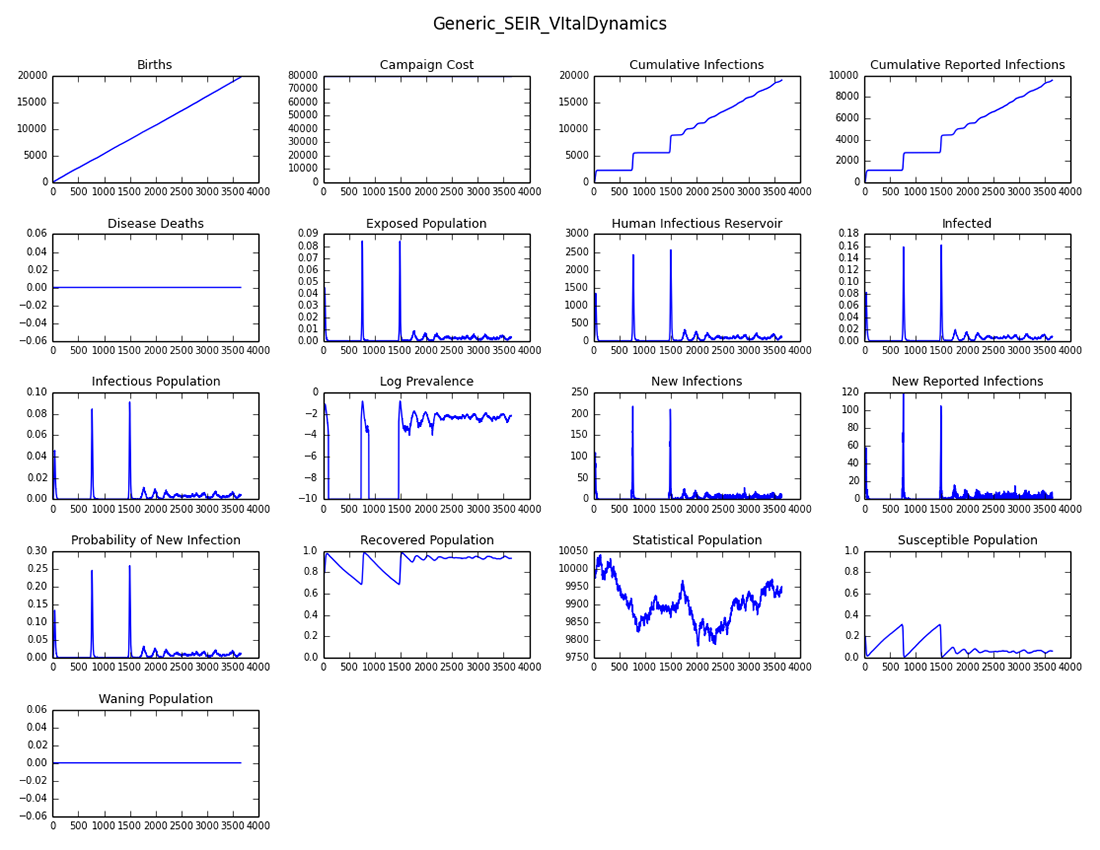
Figure 2: Periodic outbreaks on reintroduction - all channels
Exploring the model¶
Shut down the vital dynamics by setting Enable_Vital_Dynamics to 0 in the ScenariosGeneric04_SEIR_VitalDynamicsconfig.json file. Eventually, the number of susceptible individuals will be exhausted and a reintroduction of the outbreak will not cause a new outbreak.
Experiment with increasing the birth rate and population size in the generic_scenarios_demographics file. At a certain point, the influx of population might be enough to sustain the dynamics without reintroducing an outbreak.