Glossary¶
The Epidemiological MODeling software (EMOD) glossary is divided into the following subsections that define terms related to software usage, general epidemiology, and the particular disease being modeled.
Contents
Software terms¶
The following terms are used to describe both general computing processes and concepts and the files, features, and functionality related to running simulations with Epidemiological MODeling software (EMOD).
- agent-based model
- A type of simulation that models the actions and interactions of autonomous agents (both individual and collective entities such as organizations or groups).
- Boost
- Free, peer-reviewed, portable C++ source libraries aimed at a wide range of uses including parallel processing applications (Boost.MPI). For more information, please see the Boost website, http://www.boost.org.
- boxcar function
- A mathematical function that is equal to zero over the entire real line except for a single interval where it is equal to a constant.
- campaign
- A collection of events that use interventions to modify a simulation.
- campaign event
- A JSON object that determines when and where an intervention is distributed during a campaign.
- campaign file
- A JavaScript Object Notation (JSON) formatted file that contains the parameters that specify the distribution instructions for all interventions used in a campaign, such as diagnostic tests, the target demographic, and the timing and cost of interventions. The location of this file is specified in the configuration file with the Campaign_Filename parameter. Typically, the file name is campaign.json.
- channel
- A property of the simulation (for example, “Parasite Prevalence”) that is accumulated once per simulated time step and written to file, typically as an array of the accumulated values.
- class factory
- A function that instantiate objects at run-time and use information from JSON-based configuration information in the creation of these objects.
- configuration file
- A JavaScript Object Notation (JSON) formatted file that contains the parameters sufficient for initiating a simulation. It controls many different aspects of the simulation, such as population size, disease dynamics, and length of the simulation. Typically, the file name is config.json.
- core
- In computing, a core refers to an independent central processing unit (CPU) in the computer. Multi-core computers have more than one CPU. However, through technologies such as Hyper- Threading Technology (HTT or HT), a single physical core can actually act like two virtual or logical cores, and appear to the operating system as two processors.
- demographics file
- A JavaScript Object Notation (JSON) formatted file that contains the parameters that specify the demographics of a population, such as age distribution, risk, birthrate, and more. IDM provides demographics files for many geographic regions. This file is considered part of the input data files and is typically named <region>_demographics.json.
- disease-specific build
- A build of the EMOD executable (Eradication.exe) built using SCons without any dynamic link libraries (DLLs).
- dynamic link library (DLL)
- Microsoft’s implementation of a shared library, separate from the EMOD executable (Eradication.exe), that can be dynamically loaded (and unloaded when unneeded) at runtime. This loading can be explicit or implicit.
- EMODule
- A modular component of EMOD that are consumed and used by the EMOD executable (Eradication.exe). Under Windows, a EMODule is implemented as a dynamic link library (DLL) and, under CentOS on Azure, EMODules are currently not supported. EMODules are primarily custom reporters.
- Epidemiological MODeling software (EMOD)
- The modeling software from the Institute for Disease Modeling (IDM) for disease researchers and developers to investigate disease dynamics, and to assist in combating a host of infectious diseases. You may see this referred to as Disease Transmission Kernel (DTK) in the source code.
- Eradication.exe
- Typical (default) name for the EMOD executable (Eradication.exe), whether built using monolithic build or modular (EMODule-enabled) build.
- event coordinator
- A JSON object that determines who will receive a particular intervention during a campaign.
- flattened file
- A single campaign or configuration file created by combining a default file with one or more overlay files. Multiple files must be flattened prior to running a simulation. Configuration files are flattened to a single-depth JSON file without nesting, the format required for consumption by the EMOD executable (Eradication.exe). Separating the parameters into multiple files is primarily used for testing and experimentation.
- Heterogeneous Intra-Node Transmission (HINT)
- A feature for modeling person-to-person transmission of diseases in heterogeneous population segments within a node for generic simulations.
- high-performance computing (HPC)
- The use of parallel processing for running advanced applications efficiently, reliably, and quickly.
- input data files
- Fixed information used as input to a simulation. Examples of such data are population, altitude, demographics, and transportation migrations. The configurable config.json and campaign.json files do not fall into this category of static fixed input.
- intervention
- An object aimed at reducing the spread of a disease that is distributed either to an individual; such as a vaccine, drug, or bednet; or to a node; such as a larvicide. Additionally, initial disease outbreaks and intermediate interventions that schedule another intervention are implemented as interventions in the campaign file.
- JavaScript Object Notation (JSON)
- A human-readable, open standard, text-based file format for data interchange. It is typically used to represent simple data structures and associative arrays, and is language- independent.
- Keyhole Markup Language (KML)
- A file format used to display geographic data in an Earth browser, for example, Google Maps. The format uses an XML-based structure (tag-based structure with nested elements and attributes). Tags are case-sensitive.
- Link-Time Code Generation (LTCG)
- A method for the linker to optimize code (for size and/or speed) after compilation has occurred. The compiled code is turned not into actual code, but instead into an intermediate language form (IL, but not to be confused with .NET IL which has a different purpose). The LTCG then, unlike the compiler, can see the whole body of code in all object files and be able to optimize the result more effectively.
- Message Passing Interface (MPI)
- An interface used to pass information between computing cores in parallel simulations. One example is the migration of individuals from one geographic location to another within EMOD simulations.
- microsolver
- A microsolver is a type of “miniature model” that operates within the framework of EMOD to compute a particular set of parameters. Each microsolver, in effect, is creating a microsimulation in order to accurately capture the dynamics of that particular aspect of the model.
- Monte Carlo
- Monte Carlo methods, or Monte Carlo experiments, are a class of computational algorithms utilizing repeated random sampling to obtain numerical results. Monte Carlo simulations create probability distributions for possible outcome values, which provides a more realistic way of describing uncertainty in variables.
- monolithic build
- A single EMOD executable (Eradication.exe) with no DLLs that includes all components as part of Eradication.exe itself. You can still use EMODules with the monolithic build; for example, a custom reporter is a common type of EMODule. View the documentation on EMODules and emodules_map.json for more information about creation and use of EMODules.
- node
- A grid size that is used for modeling geographies. Within EMOD, a node is a geographic region containing simulated individuals. Individuals migrate between nodes either temporarily or permanently using mobility patterns driven by local, regional, and long- distance transportation.
- node-targeted intervention
- An intervention that is distributed to a geographical node rather than to a single individual. One example is larvicides, which affect all mosquitoes living and feeding within a given node.
- output report
- A file that is the output from an EMOD simulation. Output reports are in JSON, CSV, or binary file format. You must pass the data from an output report to graphing software if you want to visualize the output of a simulation.
- overlay file
- An additional configuration, campaign, or demographic file that overrides the default parameter values in the primary file. Separating the parameters into multiple files is primarily used for testing a nd experimentation. In the case of configuration and campaign files, the files can use an arbitrary hierarchical structure to organize parameters into logical groups. Configuration and campaign files must be flattened into a single file before running a simulation.
- preview
- Software that undergoes a shorter testing cycle in order to make it available more quickly. Previews may contain software defects that could result in unexpected behavior. Use EMOD previews at your own discretion.
- regression test
- A test to verify that existing EMOD functionality works with new updates, typically used to refer to one of a set of scenarios included in the EMOD bundle and located in the Regression subdirectory of the bundle. Directory names of each subdirectory in Regression describe the main regression attributes, for example, “1_Generic_Seattle_MultiNode”. Also can refer to the process of regression testing of software.
- release
- Software that includes new functionality, scientific tutorials leveraging new or existing functionality, and/or bug fixes that have been thoroughly tested so that any defects have been fixed before release. EMOD releases undergo full regression testing.
- reporter
- Functionality that extracts simulation data, aggregates it, and saves it as an output report. EMOD provides several built-in reporters for outputting data from simulations and you also have the ability to create a custom reporter.
- scenario
- A collection of input, configuration, and campaign files that describes a real-world example of a disease outbreak and interventions. Many scenarios are included with EMOD source installations or are available in the Regression directory of the EMOD GitHub repository. The tutorials describe how to run simulations using these scenarios to learn more about epidemiology and disease modeling.
- schema
- A text or JSON file that can be generated from the EMOD executable (Eradication.exe) that defines all configuration and campaign parameters.
- simulation
- An EMOD software execution performed by the kernel. Each simulation has an associated set of files that control the inputs, configuration, and campaign.
- simulation type
The disease or disease class to model.
EMOD supports the following simulation types for modeling a variety of diseases:
- Generic disease (GENERIC_SIM)
- Vector-borne diseases (VECTOR_SIM)
- Malaria (MALARIA_SIM)
- Tuberculosis (TB_SIM)
- Sexually transmitted infections (STI_SIM)
- HIV (HIV_SIM)
- solvers
- Solvers are used to find computational solutions to problems. In simulations, they can be used, for example, to determine the time of the next simulation step, or to compute the states of a model at particular time steps.
- Standard Template Library (STL)
- A library that contains a set of common C++ classes (including generic algorithms and data structures) that are independent of container and implemented as templates, which enables compile-time polymorphism (often more efficient than run-time polymorphism). For more information and discussion of STL, see Wikipedia.
- state transition event
- A change in state (e.g. healthy to infected, undiagnosed to positive diagnosis, or birth) that may trigger a subsequent action, often an intervention. “Campaign events” should not be confused with state transition events.
- time step
- A discrete number of hours or days in which the “simulation states” of all “simulation objects” (interventions, infections, immune systems, or individuals) are updated in a simulation. Each time step will complete processing before launching the next one. For example, a time step would process the migration data for populations moving between nodes via rail, airline, and road. The migration of individuals between nodes is the last step of the time step after updating states.
- tutorial
- A set of instructions in the documentation to learn more about epidemiology and disease modeling. Tutorials are based on real-world scenarios and demonstrate the mechanics of the the model. Each tutorial consists of one or more scenarios.
- working directory
- The directory that contains the configuration and campaign files for a simulation. You must be in this directory when you invoke Eradication.exe at the command line to run a simulation.
Epidemiology terms¶
The following terms are used to describe general concepts and processes in the field of epidemiology and disease modeling.
- antigen
- A substance that is capable of inducing a specific immune response and that evokes the production of one or more antibodies.
- Clausius-Clayperon relation
- A way of characterizing a transition between two phases of matter; provides a method to find a relationship between temperature and pressure along phase boundaries. Frequently used in meteorology and climatology to describe the behavior of water vapor. See Wikipedia - Clausius-Clayperon relation for more information.
- deterministic
- Characterized by the output being fully determined by the parameter values and the initial conditions. Given the same inputs, a deterministic model will always produce the same output.
- diffusive migration
- The diffusion of people in and out of nearby nodes by foot travel.
- epitope
- The portion of an antigen that the immune system recognizes. An epitope is also called an antigenic determinant.
- Euler method
- Used in mathematics and computational science, this method is a first-order numerical procedure for solving ordinary differential equations with a given initial value.
- exp(
- The exponential function,
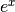 , where
, where  is the number (approximately 2.718281828)
such that the function
is the number (approximately 2.718281828)
such that the function 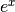 is its own derivative. The exponential function is used to
model a relationship in which a constant change in the independent variable gives the same
proportional change (i.e. percentage increase or decrease) in the dependent variable. The
function is often written as
is its own derivative. The exponential function is used to
model a relationship in which a constant change in the independent variable gives the same
proportional change (i.e. percentage increase or decrease) in the dependent variable. The
function is often written as 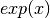 . The graph of
. The graph of 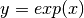 is upward-sloping
and increases faster as
is upward-sloping
and increases faster as 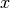 increases.
increases. - herd immunity
- The resistance to the spread of a contagious disease within a population that results if a sufficiently high proportion of individuals are immune to the disease, especially through vaccination.
- incidence
- The rate of new cases of a disease during a specified time period. This is a measure of the risk of contracting a disease.
- Koppen-Geiger Climate Classification System
- A system based on the concept that native vegetation is a good expression of climate. Thus, climate zone boundaries have been selected with vegetation distribution in mind. It combines average annual and monthly temperatures and precipitation, and the seasonality of precipitation. EMOD has several options for configuring the climate, namely air temperature, rainfall, and humidity. One option utilizes Input Files that associate geographic nodes with Koppen climate indices. The modified Koppen classification uses three letters to divide the world into five major climate regions (A, B, C, D, and E) based on average annual precipitation, average monthly precipitation, and average monthly temperature. Each category is further divided into sub-categories based on temperature and precipitation. While the Koppen system does not take such things as temperature extremes, average cloud cover, number of days with sunshine, or wind into account, it is a good representation of our earth’s climate.
- loss to follow-up (LTFU)
- Patients who at one point were actively participating in disease treatment or clinical research, but have become lost either by error or by becoming unreachable at the point of follow-up.
- ordinary differential equation (ODE)
- A differential equation containing one or more functions of one independent variable and its derivatives.
- prevalence
- The rate of all cases of a disease during a specified time period. This is a measure of how widespread a disease is.
- reproductive number
- In a fully susceptible population, the basic reproductive number R:sub:0is the number of new cases generated by one infectious case over the course of the infectious period. The effective reproductive number takes into account non-susceptible individuals.
- routine immunization (RI)
- The standard practice of vaccinating the majority of susceptible people in a population against vaccine-preventable diseases.
- scenario
- A real-world or hypothetical situation that demonstrates or represents disease dynamics.
- stochastic
- Characterized by having a random probability distribution that may be analyzed statistically but not predicted precisely.
- subpatent
- When an individual is infected but asymptomatic, so the infection is not readily detectable.
- SEIR model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible, exposed, infectious, and recovered.
- SEIRS model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible, exposed, infectious, recovered, and susceptible.
- SI model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible and infectious.
- SIR model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible, infectious, and recovered.
- SIRS model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible, infectious, recovered, and susceptible.
- SIS model
- A generic epidemiological model that provides a simplified means of describing the transmission of an infectious disease through individuals where those individuals can pass through the following five states: susceptible, infectious, and susceptible.
- supplemental immunization activity (SIA)
- In contrast to routine immunization (RI), SIAs are large-scale operations with a goal of delivering vaccines to every household.
- WAIFW matrix
- A matrix of values that describes the rate of transmission between different population groups. WAIFW is an abbrevation for Who Acquires Infection From Whom.
- Weibull distribution
- A probability distribution often used in EMOD and that requires both a shape parameter and a scale parameter. The shape parameter governs the shape of the density function. When the shape parameter is equal to 1, it is an exponential distribution. For shape parameters above 1, it forms a unimodal (hump-shaped) density function. As the shape parameter becomes large, the function forms a sharp peak. The inverse of the shape parameter is sometimes referred to here as the “heterogeneity” of the distribution (heterogeneity = 1/shape), because it can be helpful to think about the degree of heterogeneity of draws from the distribution, especially for hump-shaped functions with heterogeneity values between 0 and 1 (i.e., shape parameters greater than 1). The scale parameter shifts the distribution from left to right. When heterogeneity is small (i.e., the shape parameter is large), the scale parameter sets the location of the sharp peak.
HIV terms¶
The following terms are used to describe the biological processes involved in HIV and other sexually-transmitted infectious diseases.
- coital dilution
- The reduction in the number of sexual acts per partner when an additional, concurrent partner is added. Concurrency is defined as having two or more sexual partnerships overlapping in time.