Configuration parameters¶
The parameters described in this reference section can be added to the JavaScript Object Notation (JSON) formatted configuration file to determine the core behavior of a simulation including the computing environment, functionality to enable, additional files to use, and characteristics of the disease being modeled. This file contains mostly a flat list of JSON key:value pairs. For more information on how to use these files, see Create a configuration file.
The tables below contain only parameters available when using the HIV simulation type. Some parameters may appear in multiple categories.
Note
Parameters are case-sensitive. For Boolean parameters, set to 1 for true or 0 for false. JSON does not permit comments, but you can add “dummy” parameters to add contextual information to your files.
Contents
- Disease progression
- Drugs and treatments
- Enable or disable features
- General disease
- Geography and environment
- Immunity
- Incubation
- Infectivity and transmission
- Input data files
- Migration
- Mortality and survival
- Output settings
- Population dynamics
- Relationships and pair formation
- Sampling
- Scalars and multipliers
- Simulation setup
- Symptoms and diagnosis
- Weibull distributions
Disease progression¶
The following parameters determine aspects of HIV progression from the acute stage to AIDS.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Acute_Duration_In_Months | float | 0 | 5 | 2.9 | The time since infection, in months, over which the Acute_Stage_Infectivity_Multiplier is applied to coital acts occurring in that time period. | {
"Acute_Duration_In_Months": 2.9
}
|
| AIDS_Duration_In_Months | float | 7 | 12 | 9 | The length of time, in months, prior to an AIDS-related death over which the AIDS_Stage_Infectivity_Multiplier is applied. | {
"AIDS_Duration_In_Months": 8
}
|
Drugs and treatments¶
The following parameters determine the efficacy of drugs and other treatments.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| ART_CD4_at_Initiation_Saturating_Reduction_in_Mortality | float | 0 | 3.40E+38 | 350 | The duration from ART enrollment to on-ART HIV-caused death increases with CD4 at ART initiation up to a threshold determined by this parameter value. | {
"ART_CD4_at_Initiation_Saturating_Reduction_in_Mortality": 350
}
|
| Maternal_Transmission_ART_Multiplier | float | 0 | 1 | 0.1 | The maternal transmission multiplier for on-ART mothers. | {
"Maternal_Transmission_ART_Multiplier": 0.03
}
|
| Report_HIV_ByAgeAndGender_Collect_Intervention_Data | array of strings | NA | NA | NA | Stratifies the population by adding a column of 0s and 1s depending on whether or not the person has the indicated intervention. This only works for interventions that remain with a person for a period of time, such as ART, VMMC, vaccine/PrEP, or a delay state in the cascade of care. You can name the intervention by adding a parameter Intervention_Name in the campaign file, and then give this parameter the same Intervention_Name. This allows you to have multiple types of vaccines, VMMCs, etc., but to only stratify on the type you want. | {
"Report_HIV_ByAgeAndGender_Collect_Intervention_Data": [
"ART_Intervention",
"PrEP_Intervention"
]
}
|
Enable or disable features¶
The following parameters enable or disable features of the model, such as allowing births, deaths, or aging. Set to false (0) to disable; set to true (1) to enable.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Enable_Aging | boolean | 0 | 1 | 1 | Controls whether or not individuals in a population age during the simulation. Enable_Vital_Dynamics must be set to true (1). | {
"Enable_Vital_Dynamics": 1,
"Enable_Aging": 1
}
|
| Enable_Air_Migration | boolean | 0 | 1 | 0 | Controls whether or not migration by air travel will occur. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration": 1,
"Air_Migration_Filename": "../inputs/air_migration.bin"
}
|
| Enable_Birth | boolean | 0 | 1 | 1 | Controls whether or not individuals will be added to the simulation by birth. Enable_Vital_Dynamics must be set to true (1). If you want new individuals to have the same intervention coverage as existing individuals, you must add a BirthTriggeredIV to the campaign file. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1
}
|
| Enable_Climate_Stochasticity | boolean | 0 | 1 | 0 | Controls whether or not the climate has stochasticity. Climate_Model must be set to CLIMATE_CONSTANT or CLIMATE_BY_DATA. Set the variance using the parameters Air_Temperature_Variance, Land_Temperature_Variance, Enable_Rainfall_Stochasticity, and Relative_Humidity_Variance. | {
"Climate_Model": "CLIMATE_BY_DATA",
"Enable_Climate_Stochasticity": 1,
"Air_Temperature_Variance": 2,
"Enable_Rainfall_Stochasticity": 1,
"Land_Temperature_Variance": 2,
"Relative_Humidity_Variance": 0.05
}
|
| Enable_Coital_Dilution | boolean | 0 | 1 | 1 | Controls whether or not coital dilution will occur. | {
"Enable_Coital_Dilution": 1
}
|
| Enable_Default_Reporting | boolean | 0 | 1 | 1 | Controls whether or not the default InsetChart.json report is created. | {
"Enable_Default_Reporting": 1
}
|
| Enable_Demographics_Birth | boolean | 0 | 1 | 0 | Controls whether or not newborns have identical or heterogeneous characteristics. Set to false (0) to give all newborns identical characteristics; set to true (1) to allow for heterogeneity in traits such as sickle-cell status. Enable_Birth must be set to true (1). | {
"Enable_Birth": 1,
"Enable_Demographics_Birth": 1
}
|
| Enable_Demographics_Builtin | boolean | 0 | 1 | 0 | Controls whether or not built-in demographics for default geography will be used. Note that the built-in demographics feature does not represent a real geographical location and is mostly used for testing. Set to true (1) to define the initial population and number of nodes using Default_Geography_Initial_Node_Population and Default_Geography_Torus_Size. Set to false (0) to use demographics input files defined in Demographics_Filenames. | {
"Enable_Demographics_Builtin": 1,
"Default_Geography_Initial_Node_Population": 1000,
"Default_Geography_Torus_Size": 3
}
|
| Enable_Demographics_Gender | boolean | 0 | 1 | 1 | Controls whether or not gender ratios are drawn from a Gaussian or 50/50 draw. Set to true (1) to create gender ratios drawn from a male/female ratio that is randomly smeared by a Gaussian of width 1%; set to false (0) to assign a gender ratio based on a 50/50 draw. | {
"Enable_Demographics_Gender": 1
}
|
| Enable_Demographics_Other | boolean | 0 | 1 | 0 | Controls whether or not other demographic factors are included in the simulation, such as the fraction of individuals above poverty, urban/rural characteristics, heterogeneous initial immunity, or risk. These factors are set in the demographics file. | {
"Enable_Demographics_Other": 1
}
|
| Enable_Demographics_Reporting | boolean | 0 | 1 | 1 | Controls whether or not demographic summary data and age-binned reports are outputted to file. | {
"Enable_Demographics_Reporting": 1
}
|
| Enable_Disease_Mortality | boolean | 0 | 1 | 1 | Controls whether or not individuals die due to disease. | {
"Enable_Disease_Mortality": 1
}
|
| Enable_Family_Migration | boolean | 0 | 1 | 0 | Controls whether or not all members of a household can migrate together. All residents must be home before they can leave on the trip. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Enable_Migration": "FIXED_RATE_MIGRATION",
"Enable_Family_Migration": 1,
"Family_Migration_Filename": "../inputs/family_migration.bin"
}
|
| Enable_Heterogeneous_Intranode_Transmission | boolean | 0 | 1 | 0 | Controls whether or not individuals experience heterogeneous disease transmission within a node. When set to true (1), individual property definitions and the This is used only in generic simulations, but must be set to false (0) for all other simulation types. Heterogeneous transmission for other diseases uses other mechanistic parameters included with the simulation type. |
{
"Enable_Heterogeneous_Intranode_Transmission": 1
}
|
| Enable_Immune_Decay | boolean | 0 | 1 | 1 | Controls whether or not immunity decays after an infection clears. Set to true (1) if immunity decays; set to false (0) if recovery from the disease confers complete immunity for life. Enable_Immunity must be set to true (1). | {
"Enable_Immunity": 1,
"Enable_Immune_Decay": 0
}
|
| Enable_Immunity | boolean | 0 | 1 | 1 | Controls whether or not an individual has protective immunity after an infection clears. | {
"Enable_Immunity": 1
}
|
| Enable_Interventions | boolean | 0 | 1 | 0 | Controls whether or not campaign interventions will be used in the simulation. Set Campaign_Filename to the path of the file that contains the campaign interventions. | {
"Enable_Interventions": 1,
"Campaign_Filename": "campaign.json"
}
|
| Enable_Local_Migration | boolean | 0 | 1 | 0 | Controls whether or not local migration (the diffusion of people in and out of nearby nodes by foot travel) occurs. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Local_Migration": 1,
"Local_Migration_Filename": "../inputs/local_migration.bin"
}
|
| Enable_Maternal_Transmission | boolean | 0 | 1 | 0 | Controls whether or not infectious mothers infect infants at birth. Enable_Birth must be set to true (1). | {
"Enable_Birth": 1,
"Enable_Maternal_Transmission": 1
}
|
| Enable_Migration_Heterogeneity | boolean | 0 | 1 | 1 | Controls whether or not migration rate is heterogeneous among individuals. Set to true (1) to use a migration rate distribution in the demographics file (see NodeAttributes parameters); set to false (0) to use the same migration rate applied to all individuals. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Migration_Heterogeneity": 1
}
|
| Enable_Property_Output | boolean | 0 | 1 | 0 | Controls whether or not to create property output reports, which detail groups as defined in IndividualProperties in the demographics file (see NodeProperties and IndividualProperties parameters). When there is more than one property type, the report will display the channel information for all combinations of the property type groups. | {
"Enable_Property_Output": 1
}
|
| Enable_Rainfall_Stochasticity | boolean | 0 | 1 | 1 | Controls whether or not there is stochastic variation in rainfall; set to true (1) for stochastic variation of rainfall that is drawn from an exponential distribution (with a mean value as the daily rainfall from the Climate_Model values CLIMATE_CONSTANT or CLIMATE_BY_DATA), or set to false (0) to disable rainfall stochasticity. | {
"Enable_Climate_Stochasticity": 1,
"Air_Temperature_Variance": 2,
"Enable_Rainfall_Stochasticity": 1,
"Land_Temperature_Variance": 2,
"Relative_Humidity_Variance": 0.05
}
|
| Enable_Regional_Migration | boolean | 0 | 1 | 0 | Controls whether or not there is migration by road vehicle into and out of nodes in the road network. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Regional_Migration": 1,
"Regional_Migration_Filename": "../inputs/regional_migration.bin"
}
|
| Enable_Sea_Migration | boolean | 0 | 1 | 0 | Controls whether or not there is migration on ships into and out of coastal cities with seaports. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Sea_Migration": 1,
"Sea_Migration_Filename": "../inputs/sea_migration.bin"
}
|
| Enable_Spatial_Output | boolean | 0 | 1 | 0 | Controls whether or not spatial output reports are created. If set to true (1), spatial output reports include all channels listed in the parameter Spatial_Output_Channels. Note Spatial output files require significant processing time and disk space. |
{
"Enable_Spatial_Output": 1,
"Spatial_Output_Channels": [
"Prevalence",
"New_Infections"
]
}
|
| Enable_Superinfection | boolean | 0 | 1 | 0 | Controls whether or not an individual can have multiple infections simultaneously. Set to true (1) to allow for multiple simultaneous infections; set to false (0) if multiple infections are not possible. Set the Max_Individual_Infections parameter. | {
"Enable_Superinfection": 1,
"Max_Individual_Infections": 2
}
|
| Enable_Vital_Dynamics | boolean | 0 | 1 | 1 | Controls whether or not births and deaths occur in the simulation. Births and deaths must be individually enabled and set. | {
"Enable_Vital_Dynamics":1,
"Enable_Birth": 1,
"Death_Rate_Dependence": "NONDISEASE_MORTALITY_OFF",
"Base_Mortality": 0.002
}
|
General disease¶
The following parameters determine general disease characteristics.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Animal_Reservoir_Type | enum | NA | NA | NO_ZOONOSIS | The type of animal reservoir and configuration for zoonosis. Use of the animal reservoir sets a low constant baseline of infectivity beyond what is present in the human population. It allows a more random introduction of cases in continuous time, which is more applicable for various situations such as zoonosis. Possible values are:
|
{
"Animal_Reservoir_Type": "CONSTANT_ZOONOSIS"
}
|
| Number_Basestrains | integer | 1 | 10 | 1 | The number of base strains in the simulation, such as antigenic variants. | {
"Number_Basestrains": 1
}
|
| Number_Substrains | integer | 1 | 16777200 | 256 | The number of disease substrains for each base strain, such as genetic variants. | {
"Number_Substrains": 16777216
}
|
| Zoonosis_Rate | float | 0 | 1 | 0 | The daily rate of zoonotic infection per individual. Animal_Reservoir_Type must be set to CONSTANT_ZOONOSIS or ZOONOSIS_FROM_DEMOGRAPHICS. If Animal_Reservoir_Type is set to ZOONOSIS_FROM_DEMOGRAPHICS, the value for the Zoonosis NodeAttribute in the demographics file will override the value set for Zoonosis_Rate. | {
"Zoonosis_Rate": 0.005
}
|
Geography and environment¶
The following parameters determine characteristics of the geography and environment of the simulation. For example, how to use the temperature or rainfall data in the climate files and the size of the nodes in the simulation.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Air_Temperature_Filename | string | NA | NA | air_temp.json | The path to the input data file that defines air temperature data measured two meters above ground. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Climate_Model": "CLIMATE_BY_DATA",
"Air_Temperature_Filename": "Namawala_single_node_air_temperature_daily.bin"
}
|
| Air_Temperature_Offset | float | -20 | 20 | 0 | The linear shift of air temperature in degrees Celsius. Climate_Model must be set to CLIMATE_BY_DATA. | {
"Air_Temperature_Offset": 1
}
|
| Air_Temperature_Variance | float | 0 | 5 | 2 | The standard deviation (in degrees Celsius) for normally distributed noise applied to the daily air temperature values when Climate_Model is configured as CLIMATE_CONSTANT or CLIMATE_BY_DATA. Enable_Climate_Stochasticity must be set to true (1). | {
"Enable_Climate_Stochasticity": 1,
"Air_Temperature_Variance": 2,
}
|
| Base_Air_Temperature | float | -55 | 45 | 22 | The air temperature, in degrees Celsius, where Climate_Model is set to CLIMATE_CONSTANT. | {
"Climate_Model": "CLIMATE_CONSTANT",
"Base_Air_Temperature": 30
}
|
| Base_Land_Temperature | float | -55 | 60 | 26 | The land temperature, in degrees Celsius, where Climate_Model is set to CLIMATE_CONSTANT. | {
"Climate_Model": "CLIMATE_CONSTANT",
"Base_Land_Temperature": 20
}
|
| Base_Rainfall | float | 0 | 150 | 10 | The value of rainfall per day in millimeters when Climate_Model is set to CLIMATE_CONSTANT. | {
"Climate_Model": "CLIMATE_CONSTANT",
"Base_Rainfall": 20
}
|
| Base_Relative_Humidity | float | 0 | 1 | 0.75 | The value of humidity where Climate_Model is set to CLIMATE_CONSTANT. | {
"Base_Relative_Humidity": 0.1
}
|
| Climate_Model | enum | NA | NA | CLIMATE_OFF | How and from what files the climate of a simulation is configured. The possible values are:
|
{
"Climate_Model": "CLIMATE_CONSTANT"
}
|
| Climate_Update_Resolution | enum | NA | NA | CLIMATE_UPDATE_YEAR | The resolution of data in climate files. Climate_Model must be set to CLIMATE_CONSTANT, CLIMATE_BY_DATA, or CLIMATE_KOPPEN. Possible values are: CLIMATE_UPDATE_YEAR CLIMATE_UPDATE_MONTH CLIMATE_UPDATE_WEEK CLIMATE_UPDATE_DAY CLIMATE_UPDATE_HOUR |
{
"Climate_Update_Resolution": "CLIMATE_UPDATE_DAY"
}
|
| Default_Geography_Initial_Node_Population | integer | 0 | 1000000 | 1000 | When using the built-in demographics for default geography, the initial number of individuals in each node. Note that the built-in demographics feature does not represent a real geographical location and is mostly used for testing. Enable_Demographics_Builtin must be set to true (1). | {
"Enable_Demographics_Builtin": 1,
"Default_Geography_Initial_Node_Population": 1000,
"Default_Geography_Torus_Size": 3
}
|
| Default_Geography_Torus_Size | integer | 3 | 100 | 10 | When using the built-in demographics for default geography, the square root of the number of nodes in the simulation. The simulation uses an N x N square grid of nodes with N specified by this parameter. If migration is enabled, the N x N nodes are assumed to be a torus and individuals can migrate from any node to all four adjacent nodes. To enable migration, set Migration_Model to FIXED_RATE_MIGRATION. Built-in migration is a form of “local” migration where individuals only migrate to the adjacent nodes. You can use the x_Local_Migration parameter to control the rate of migration. The other migration parameters are ignored. Note that the built-in demographics feature does not represent a real geographical location and is mostly used for testing. Enable_Demographics_Builtin must be set to true (1). |
{
"Enable_Demographics_Builtin": 1,
"Default_Geography_Initial_Node_Population": 1000,
"Default_Geography_Torus_Size": 3
}
|
| Enable_Climate_Stochasticity | boolean | 0 | 1 | 0 | Controls whether or not the climate has stochasticity. Climate_Model must be set to CLIMATE_CONSTANT or CLIMATE_BY_DATA. Set the variance using the parameters Air_Temperature_Variance, Land_Temperature_Variance, Enable_Rainfall_Stochasticity, and Relative_Humidity_Variance. | {
"Climate_Model": "CLIMATE_BY_DATA",
"Enable_Climate_Stochasticity": 1,
"Air_Temperature_Variance": 2,
"Enable_Rainfall_Stochasticity": 1,
"Land_Temperature_Variance": 2,
"Relative_Humidity_Variance": 0.05
}
|
| Enable_Rainfall_Stochasticity | boolean | 0 | 1 | 1 | Controls whether or not there is stochastic variation in rainfall; set to true (1) for stochastic variation of rainfall that is drawn from an exponential distribution (with a mean value as the daily rainfall from the Climate_Model values CLIMATE_CONSTANT or CLIMATE_BY_DATA), or set to false (0) to disable rainfall stochasticity. | {
"Enable_Climate_Stochasticity": 1,
"Air_Temperature_Variance": 2,
"Enable_Rainfall_Stochasticity": 1,
"Land_Temperature_Variance": 2,
"Relative_Humidity_Variance": 0.05
}
|
| Koppen_Filename | string | NA | NA | UNINITIALIZED STRING | The path to the input file used to specify Koppen climate classifications; only used when Climate_Model is set to CLIMATE_KOPPEN. The file must be in .dat format. | {
"Koppen_Filename": "Mad_2_5arcminute_koppen.dat"
}
|
| Land_Temperature_Filename | string | NA | NA | land_temp.json | The path of the input file defining temperature data measured at land surface; used only when Climate_Model is set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Land_Temperature_Filename": "Namawala_single_node_land_temperature_daily.bin"
}
|
| Land_Temperature_Offset | float | -20 | 20 | 0 | The linear shift of land surface temperature in degrees Celsius; only used when Climate_Model is set to CLIMATE_BY_DATA. | {
"Land_Temperature_Offset": 0
}
|
| Land_Temperature_Variance | float | 0 | 7 | 2 | The standard deviation (in degrees Celsius) for normally distributed noise applied to the daily land temperature values when Climate_Model is configured to CLIMATE_CONSTANT or CLIMATE_BY_DATA; only used if the Enable_Climate_Stochasticity is set to true (1). | {
"Land_Temperature_Variance": 1.5
}
|
| Node_Grid_Size | float | 0.00416 | 90 | 0.004167 | The spatial resolution indicating the node grid size for a simulation in degrees. | {
"Node_Grid_Size": 0.042
}
|
| Rainfall_Filename | string | NA | NA | rainfall.json | The path of the input file which defines rainfall data. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Rainfall_Filename": "Namawala_single_node_rainfall_daily.bin"
}
|
| Rainfall_Scale_Factor | float | 0.1 | 10 | 1 | The scale factor used in multiplying rainfall value(s). Climate_Model must be set to CLIMATE_BY_DATA. | {
"Rainfall_Scale_Factor": 1
}
|
| Relative_Humidity_Filename | string | NA | NA | rel_hum.json | The path of the input file which defines relative humidity data measured 2 meters above ground. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Relative_Humidity_Filename": "Namawala_single_node_relative_humidity_daily.bin"
}
|
| Relative_Humidity_Scale_Factor | float | 0.1 | 10 | 1 | The scale factor used in multiplying relative humidity values. Climate_Model must be set to CLIMATE_BY_DATA. | {
"Relative_Humidity_Scale_Factor": 1
}
|
| Relative_Humidity_Variance | float | 0 | 0.12 | 0.05 | The standard deviation (in percentage) for normally distributed noise applied to the daily relative humidity values when Climate_Model is configured as CLIMATE_CONSTANT or CLIMATE_BY_DATA. Enable_Climate_Stochasticity must be set to true (1). | {
"Relative_Humidity_Variance": 0.05
}
|
Immunity¶
The following parameters determine the immune system response for the disease being modeled, including waning immunity after an infection clears.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Acquisition_Blocking_Immunity_Decay_Rate | float | 0 | 1000 | 0.001 | The rate at which acquisition-blocking immunity decays after the initial period indicated by the base acquisition-blocking immunity offset. Only used when Enable_Immunity and Enable_Immune_Decay parameters are set to true (1). | {
"Acquisition_Blocking_Immunity_Decay_Rate": 0.05
}
|
| Acquisition_Blocking_Immunity_Duration_Before_Decay | float | 0 | 45000 | 0 | The number of days after infection until acquisition-blocking immunity begins to decay. Enable_Immunity and Enable_Immune_Decay must be set to true (1). | {
"Acquisition_Blocking_Immunity_Duration_Before_Decay": 10
}
|
| Enable_Immune_Decay | boolean | 0 | 1 | 1 | Controls whether or not immunity decays after an infection clears. Set to true (1) if immunity decays; set to false (0) if recovery from the disease confers complete immunity for life. Enable_Immunity must be set to true (1). | {
"Enable_Immunity": 1,
"Enable_Immune_Decay": 0
}
|
| Enable_Immunity | boolean | 0 | 1 | 1 | Controls whether or not an individual has protective immunity after an infection clears. | {
"Enable_Immunity": 1
}
|
| Immune_Threshold_For_Downsampling | float | 0 | 1 | 0 | Threshold on acquisition immunity at which to apply immunity dependent downsampling. Individual_Sampling_Type must set to ADAPTED_SAMPLING_BY_IMMUNE_STATE. | {
"Individual_Sampling_Type": "ADAPTED_SAMPLING_BY_IMMUNE_STATE",
"Immune_Threshold_For_Downsampling": 0.5
}
|
| Immunity_Acquisition_Factor | float | 0 | 1000 | 0 | The multiplicative reduction in the probability of reacquiring disease. Only used when Enable_Immunity and Enable_Immune_Decay are set to 1. | {
"Enable_Immunity": 1,
"Enable_Immune_Decay": 1,
"Immunity_Acquisition_Factor": 0.9
}
|
| Immunity_Initialization_Distribution_Type | enum | NA | NA | DISTRIBUTION_OFF | The method for initializing the immunity distribution in the simulated population. Enable_Immunity must be set to true (1). Possible values are:
|
{
"Immunity_Initialization_Distribution_Type": "DISTRIBUTION_COMPLEX"
}
|
| Immunity_Mortality_Factor | float | 0 | 1000 | 0 | The multiplicative reduction in the probability of dying from infection after getting re-infected. Enable_Immunity and Enable_Immune_Decay must be set to true (1). | {
"Enable_Immunity": 1,
"Enable_Immune_Decay": 1,
"Immunity_Mortality_Factor": 0.5
}
|
| Immunity_Transmission_Factor | float | 0 | 1000 | 0 | The multiplicative reduction in the probability of transmitting infection after getting re-infected. Only used when Enable_Immunity and Enable_Immune_Decay are set to 1. | {
"Enable_Immunity": 1,
"Enable_Immunity_Decay": 1,
"Immunity_Transmission_Factor": 0.9
}
|
| Mortality_Blocking_Immunity_Decay_Rate | float | 0 | 1000 | 0.001 | The rate at which mortality-blocking immunity decays after the mortality-blocking immunity offset period. Enable_Immune_Decay must be set to 1. | {
"Mortality_Blocking_Immunity_Decay_Rate": 0.1
}
|
| Mortality_Blocking_Immunity_Duration_Before_Decay | float | 0 | 45000 | 0 | The number of days after infection until mortality-blocking immunity begins to decay. Enable_Immunity and Enable_Immune_Decay must be set to 1. | {
"Mortality_Blocking_Immunity_Duration_Before_Decay": 270
}
|
| Transmission_Blocking_Immunity_Decay_Rate | float | 0 | 1000 | 0.001 | The rate at which transmission-blocking immunity decays after the base transmission-blocking immunity offset period. Used only when Enable_Immunity and Enable_Immune_Decay parameters are set to true (1). | {
"Transmission_Blocking_Immunity_Decay_Rate": 0.01
}
|
| Transmission_Blocking_Immunity_Duration_Before_Decay | float | 0 | 45000 | 0 | The number of days after infection until transmission-blocking immunity begins to decay. Only used when Enable_Immunity and Enable_Immune_Decay parameters are set to true (1). | {
"Transmission_Blocking_Immunity_Duration_Before_Decay": 90
}
|
Incubation¶
The following parameters determine the characteristics of the incubation period.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Base_Incubation_Period | float | 0 | 3.40E+38 | 6 | Average duration, in days, of the incubation period before infected individuals become infectious. Incubation_Period_Distribution must be set to either FIXED_DURATION or EXPONENTIAL_DURATION. | {
"Incubation_Period_Distribution": "EXPONENTIAL_DURATION",
"Base_Incubation_Period": 1
}
|
| Incubation_Period_Distribution | enum | NA | NA | NOT_INITIALIZED | The distribution for the duration of the incubation period. Possible values are:
|
{
"Base_Incubation_Period": 5,
"Incubation_Period_Distribution": "EXPONENTIAL_DURATION"
}
|
| Incubation_Period_Log_Mean | float | 0 | 3.40E+38 | 6 | The mean of log normal for the incubation period distribution. Incubation_Period_Distribution must be set to LOG_NORMAL_DURATION. | {
"Incubation_Period_Distribution": "LOG_NORMAL_DURATION",
"Incubation_Period_Log_Mean": 5.758,
"Incubation_Period_Log_Width": 0.27
}
|
| Incubation_Period_Log_Width | float | 0 | 3.40E+38 | 1 | The log width of log normal for the incubation period distribution. Incubation_Period_Distribution must be set to LOG_NORMAL_DURATION. | {
"Incubation_Period_Distribution": "LOG_NORMAL_DURATION",
"Incubation_Period_Log_Mean": 5.758,
"Incubation_Period_Log_Width": 0.27
}
|
| Incubation_Period_Max | float | 0.6 | 3.40E+38 | 0 | The maximum length of the incubation period. Incubation_Period_Distribution must be set to UNIFORM_DURATION. | {
"Incubation_Period_Distribution": "UNIFORM_DURATION",
"Incubation_Period_Min": 2,
"Incubation_Period_Max": 6
}
|
| Incubation_Period_Mean | float | 0 | 3.40E+38 | 6 | The mean of the incubation period. Incubation_Period_Distribution must be set to either GAUSSIAN_DURATION or POISSON_DURATION. | {
"Incubation_Period_Distribution": "GAUSSIAN_DURATION",
"Incubation_Period_Mean": 7,
"Infectious_Period_Std_Dev": 2
}
|
| Incubation_Period_Min | float | 0 | 3.40E+38 | 0 | The minimum length of the incubation period. Incubation_Period_Distribution must be set to UNIFORM_DURATION. | {
"Incubation_Period_Distribution": "UNIFORM_DURATION",
"Incubation_Period_Min": 2,
"Incubation_Period_Max": 6
}
|
| Incubation_Period_Std_Dev | float | 0 | 3.40E+38 | 1 | The standard deviation incubation period. Incubation_Period_Distribution must be set to GAUSSIAN_DURATION. | {
"Incubation_Period_Distribution": "GAUSSIAN_DURATION",
"Incubation_Period_Mean": 7,
"Infectious_Period_Std_Dev": 2
}
|
Infectivity and transmission¶
The following parameters determine aspects of infectivity and disease transmission. For example, how infectious individuals are and the length of time for which they remain infectious, whether the disease can be maternally transmitted, and how population density affects infectivity.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Acute_Duration_In_Months | float | 0 | 5 | 2.9 | The time since infection, in months, over which the Acute_Stage_Infectivity_Multiplier is applied to coital acts occurring in that time period. | {
"Acute_Duration_In_Months": 2.9
}
|
| Acute_Stage_Infectivity_Multiplier | float | 1 | 100 | 26 | The multiplier acting on Base_Infectivity to determine the per-act transmission probability of an HIV+ individual during the acute stage. | {
"Acute_Stage_Infectivity_Multiplier": 3
}
|
| AIDS_Stage_Infectivity_Multiplier | float | 1 | 100 | 10 | The multiplier acting on Base_Infectivity to determine the per-act transmission probability of an HIV+ individual during the AIDS stage. | {
"AIDS_Stage_Infectivity_Multiplier": 8
}
|
| ART_Viral_Suppression_Multiplier | float | 0 | 1 | 0.08 | Multiplier acting on Base_Infectivity to determine the per-act transmission probability of a virally suppressed HIV+ individual. | {
"ART_Viral_Suppression_Multiplier": 0.3
}
|
| Base_Infectious_Period | float | 0 | 3.40E+38 | 6 | Average duration, in days, of the infectious period before the infection is cleared. Infectious_Period_Distribution must be set to either FIXED_DURATION or EXPONENTIAL_DURATION. | {
"Base_Infectious_Period": 4
}
|
| Base_Infectivity | float | 0 | 1000 | 0.3 | The base infectiousness of individuals before accounting for transmission-blocking effects of acquired immunity and/or campaign interventions. | {
"Base_Infectivity": 0.5
}
|
| CD4_At_Death_LogLogistic_Heterogeneity | float | 0 | 100 | 0 | The inverse shape parameter of a Weibull distribution that represents the at-death CD4 cell count. | {
"CD4_At_Death_LogLogistic_Heterogeneity": 0.7
}
|
| Condom_Transmission_Blocking_Probability | float | 0 | 1 | 0.9 | The per-act multiplier of the transmission probability when a condom is used. | {
"Condom_Transmission_Blocking_Probability": 0.99
}
|
| Enable_Heterogeneous_Intranode_Transmission | boolean | 0 | 1 | 0 | Controls whether or not individuals experience heterogeneous disease transmission within a node. When set to true (1), individual property definitions and the This is used only in generic simulations, but must be set to false (0) for all other simulation types. Heterogeneous transmission for other diseases uses other mechanistic parameters included with the simulation type. |
{
"Enable_Heterogeneous_Intranode_Transmission": 1
}
|
| Enable_Maternal_Transmission | boolean | 0 | 1 | 0 | Controls whether or not infectious mothers infect infants at birth. Enable_Birth must be set to true (1). | {
"Enable_Birth": 1,
"Enable_Maternal_Transmission": 1
}
|
| Enable_Superinfection | boolean | 0 | 1 | 0 | Controls whether or not an individual can have multiple infections simultaneously. Set to true (1) to allow for multiple simultaneous infections; set to false (0) if multiple infections are not possible. Set the Max_Individual_Infections parameter. | {
"Enable_Superinfection": 1,
"Max_Individual_Infections": 2
}
|
| Heterogeneous_Infectiousness_LogNormal_Scale | float | 0 | 10 | 0 | Scale parameter of a log normal distribution that governs an infectiousness multiplier. The multiplier represents heterogeneity in infectivity and adjusts Base_Infectivity. | {
"Heterogeneous_Infectiousness_LogNormal_Scale": 1
}
|
| Infection_Updates_Per_Timestep | integer | 0 | 144 | 1 | The number of infection updates executed during each timestep; note that a timestep defaults to one day. | {
"Infection_Updates_Per_Timestep": 1
}
|
| Infectious_Period_Distribution | enum | NA | NA | NOT_INITIALIZED | The distribution of the duration of the infectious period. Possible values are:
|
{
"Infectious_Period_Distribution": "EXPONENTIAL_DURATION"
}
|
| Infectious_Period_Max | float | 0.6 | 3.40E+38 | 0 | The maximum length of the infectious period; used when Infectious_Period_Distribution is set to UNIFORM_DURATION. | {
"Infectious_Period_Distribution": "UNIFORM_DURATION",
"Infectious_Period_Max": 15,
"Infectious_Period_Min": 5
}
|
| Infectious_Period_Mean | float | 0 | 3.40E+38 | 6 | The mean of the infectious period; used when Infectious_Period_Distribution is set to either GAUSSIAN_DURATION or POISSON_DURATION. | {
"Infectious_Period_Distribution": "GAUSSIAN_DURATION",
"Infectious_Period_Mean": 12,
"Infectious_Period_Std_Dev": 10
}
|
| Infectious_Period_Min | float | 0 | 3.40E+38 | 0 | The minimum length of the infectious period; used when Infectious_Period_Distribution is set to UNIFORM_DURATION. | {
"Infectious_Period_Distribution": "UNIFORM_DURATION",
"Infectious_Period_Max": 15,
"Infectious_Period_Min": 5
}
|
| Infectious_Period_Std_Dev | float | 0 | 3.40E+38 | 1 | The standard deviation of the infectious period; used when Infectious_Period_Distribution is set to GAUSSIAN_DURATION. | {
"Infectious_Period_Distribution": "GAUSSIAN_DURATION",
"Infectious_Period_Mean": 12,
"Infectious_Period_Std_Dev": 10
}
|
| Infectivity_Boxcar_Forcing_Amplitude | float | 0 | 3.40E+38 | 0 | The fractional increase in R0 during the high-infectivity season when Infectivity_Scale_Type is equal to ANNUAL_BOXCAR_FUNCTION. | {
"Infectivity_Boxcar_Forcing_Amplitude": 0.25,
"Infectivity_Boxcar_Forcing_End_Time": 270,
"Infectivity_Boxcar_Forcing_Start_Time": 90,
"Infectivity_Scale_Type": "ANNUAL_BOXCAR_FUNCTION"
}
|
| Infectivity_Boxcar_Forcing_End_Time | float | 0 | 365 | 0 | The end of the high-infectivity season when Infectivity_Scale_Type is equal to ANNUAL_BOXCAR_FUNCTION. | {
"Infectivity_Boxcar_Forcing_Amplitude": 0.25,
"Infectivity_Boxcar_Forcing_End_Time": 270,
"Infectivity_Boxcar_Forcing_Start_Time": 90,
"Infectivity_Scale_Type": "ANNUAL_BOXCAR_FUNCTION"
}
|
| Infectivity_Boxcar_Forcing_Start_Time | float | 0 | 365 | 0 | The beginning of the high-infectivity season, in days, when Infectivity_Scale_Type is equal to ANNUAL_BOXCAR_FUNCTION. | {
"Infectivity_Boxcar_Forcing_Amplitude": 0.25,
"Infectivity_Boxcar_Forcing_End_Time": 270,
"Infectivity_Boxcar_Forcing_Start_Time": 90,
"Infectivity_Scale_Type": "ANNUAL_BOXCAR_FUNCTION"
}
|
| Infectivity_Exponential_Baseline | float | 0 | 1 | 0 | The scale factor applied to Base_Infectivity at the beginning of a simulation, before the infectivity begins to grow exponentially. Infectivity_Scale_Type must be set to EXPONENTIAL_FUNCTION_OF_TIME. | {
"Infectivity_Exponential_Baseline": 0.1,
"Infectivity_Exponential_Delay": 90,
"Infectivity_Exponential_Rate": 45,
"Infectivity_Scale_Type": "EXPONENTIAL_FUNCTION_OF_TIME"
}
|
| Infectivity_Exponential_Delay | float | 0 | 3.40E+38 | 0 | The number of days before infectivity begins to ramp up exponentially. Infectivity_Scale_Type must be set to EXPONENTIAL_FUNCTION_OF_TIME. | {
"Infectivity_Exponential_Baseline": 0.1,
"Infectivity_Exponential_Delay": 90,
"Infectivity_Exponential_Rate": 45,
"Infectivity_Scale_Type": "EXPONENTIAL_FUNCTION_OF_TIME"
}
|
| Infectivity_Exponential_Rate | float | 0 | 3.40E+38 | 0 | The daily rate of exponential growth to approach to full infectivity after the delay set by Infectivity_Exponential_Delay has passed. Infectivity_Scale_Type must be set to EXPONENTIAL_FUNCTION_OF_TIME. | {
"Infectivity_Exponential_Rate": 45
}
|
| Infectivity_Scale_Type | enum | NA | NA | CONSTANT_INFECTIVITY | A scale factor that allows infectivity to be altered by time or season. Possible values are:
|
{
"Infectivity_Scale_Type": "FUNCTION_OF_CLIMATE"
}
|
| Infectivity_Sinusoidal_Forcing_Amplitude | float | 0 | 1 | 0 | Sets the amplitude of sinusoidal variations in Base_Infectivity. Only used when Infectivity_Scale_Type is set to SINUSOIDAL_FUNCTION_OF_TIME. | {
"Infectivity_Scale_Type": "SINUSOIDAL_FUNCTION_OF_TIME",
"Infectivity_Sinusoidal_Forcing_Amplitude": 0.1,
"Infectivity_Sinusoidal_Forcing_Phase": 0
}
|
| Infectivity_Sinusoidal_Forcing_Phase | float | 0 | 365 | 0 | Sets the phase of sinusoidal variations in Base_Infectivity. Only used when Infectivity_Scale_Type is set to SINUSOIDAL_FUNCTION_OF_TIME. | {
"Infectivity_Scale_Type": "SINUSOIDAL_FUNCTION_OF_TIME",
"Infectivity_Sinusoidal_Forcing_Amplitude": 0.1,
"Infectivity_Sinusoidal_Forcing_Phase": 0
}
|
| Male_To_Female_Relative_Infectivity_Ages | array of floats | NA | NA | 0 | The vector of ages governing the susceptibility of females relative to males, by age. Used with Male_To_Female_Relative_Infectivity_Multipliers. | {
"Male_To_Female_Relative_Infectivity_Ages": [
15,
25,
35
]
}
|
| Male_To_Female_Relative_Infectivity_Multipliers | array of floats | NA | NA | 1 | An array of scale factors governing the susceptibility of females relative to males, by age. Used with Male_To_Female_Relative_Infectivity_Ages. Scaling is linearly interpolated between ages. The first value is used for individuals younger than the first age in Male_To_Female_Relative_Infectivity_Ages and the last value is used for individuals older than the last age. | {
"Male_To_Female_Relative_Infectivity_Multipliers": [
5,
1,
0.5
]
}
|
| Maternal_Transmission_ART_Multiplier | float | 0 | 1 | 0.1 | The maternal transmission multiplier for on-ART mothers. | {
"Maternal_Transmission_ART_Multiplier": 0.03
}
|
| Maternal_Transmission_Probability | float | 0 | 1 | 0 | The probability of transmission of infection from mother to infant at birth. Enable_Maternal_Transmission must be set to 1. Note: For malaria and vector simulations, set this to 0. Instead, use the Maternal_Antibody_Protection, Maternal_Antibody_Decay_Rate, and Maternal_Antibodies_Type parameters. | {
"Maternal_Transmission_Probability": 0.3
}
|
| Max_Individual_Infections | integer | 0 | 1000 | 1 | The limit on the number of infections that an individual can have simultaneously. Enable_Superinfection must be set to 1. | {
"Max_Individual_Infections": 5
}
|
| Population_Density_C50 | float | 0 | 3.40E+38 | 10 | The population density at which R0 for a 2.5-arc minute square reaches half of its initial value. Population_Density_Infectivity_Correction must be set to SATURATING_FUNCTION_OF_DENSITY. | {
"Population_Density_C50": 30
}
|
| Population_Density_Infectivity_Correction | enum | NA | NA | CONSTANT_INFECTIVITY | Correction to alter infectivity by population density set in the Population_Density_C50 parameter. Measured in people per square kilometer. Possible values are:
Note Sparsely populated areas have a lower infectivity, while densely populated areas have a higher infectivity, which rises to saturate at the Base_Infectivity value. |
{
"Population_Density_Infectivity_Correction": "SATURATING_FUNCTION_OF_DENSITY"
}
|
| Relative_Sample_Rate_Immune | float | 0.001 | 1 | 0.1 | The relative sampling rate for people who have acquired immunity through recovery or vaccination. | {
"Relative_Sample_Rate_Immune": 0.1
}
|
| Susceptibility_Scale_Type | enum | NA | NA | CONSTANT_SUSCEPTIBILITY | The effect of time or season on infectivity. Possible values are: CONSTANT_SUSCEPTIBILITY LOG_LINEAR_FUNCTION_OF_TIME LINEAR_FUNCTION_OF_AGE LOG_LINEAR_FUNCTION_OF_AGE |
{
"Susceptibility_Scale_Type": "CONSTANT_SUSCEPTIBILITY"
}
|
| Transmission_Blocking_Immunity_Decay_Rate | float | 0 | 1000 | 0.001 | The rate at which transmission-blocking immunity decays after the base transmission-blocking immunity offset period. Used only when Enable_Immunity and Enable_Immune_Decay parameters are set to true (1). | {
"Transmission_Blocking_Immunity_Decay_Rate": 0.01
}
|
| Transmission_Blocking_Immunity_Duration_Before_Decay | float | 0 | 45000 | 0 | The number of days after infection until transmission-blocking immunity begins to decay. Only used when Enable_Immunity and Enable_Immune_Decay parameters are set to true (1). | {
"Transmission_Blocking_Immunity_Duration_Before_Decay": 90
}
|
| Zoonosis_Rate | float | 0 | 1 | 0 | The daily rate of zoonotic infection per individual. Animal_Reservoir_Type must be set to CONSTANT_ZOONOSIS or ZOONOSIS_FROM_DEMOGRAPHICS. If Animal_Reservoir_Type is set to ZOONOSIS_FROM_DEMOGRAPHICS, the value for the Zoonosis NodeAttribute in the demographics file will override the value set for Zoonosis_Rate. | {
"Zoonosis_Rate": 0.005
}
|
Input data files¶
The following parameters set the paths to the the campaign file and the input data files for climate, migration, demographics, and load-balancing.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Air_Migration_Filename | string | NA | NA | The path to the input data file that defines patterns of migration by airplane. Enable_Air_Migration must be set to true (1). The file must be in .bin format. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration" : 1,
"Air_Migration_Filename": "../Global_1degree_air_migration.bin"
}
|
|
| Air_Temperature_Filename | string | NA | NA | air_temp.json | The path to the input data file that defines air temperature data measured two meters above ground. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Climate_Model": "CLIMATE_BY_DATA",
"Air_Temperature_Filename": "Namawala_single_node_air_temperature_daily.bin"
}
|
| Campaign_Filename | string | NA | NA | The path to the campaign file. It is required when interventions are part of the simulation and Enable_Interventions is set to true (1). The file must be in .json format. | {
"Enable_Interventions": 1,
"Campaign_Filename": "campaign.json"
}
|
|
| Demographics_Filenames | array of strings | NA | NA | An array of the paths to demographics files containing information on the identity and demographics of the region to simulate. The files must be in .json format. | {
"Demographics_Filenames": [
"Namawala_single_node_demographics.json",
"Namawala_demographics_overlay.json"
]
}
|
|
| Family_Migration_Filename | string | NA | NA | The name of the binary file to use to configure family migration. Enable_Family_Migration must be set to true (1). The file must be in .bin format. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Family_Migration" 1,
"Family_Migration_Filename": "../inputs/family_migration.bin"
}
|
|
| Koppen_Filename | string | NA | NA | UNINITIALIZED STRING | The path to the input file used to specify Koppen climate classifications; only used when Climate_Model is set to CLIMATE_KOPPEN. The file must be in .dat format. | {
"Koppen_Filename": "Mad_2_5arcminute_koppen.dat"
}
|
| Land_Temperature_Filename | string | NA | NA | land_temp.json | The path of the input file defining temperature data measured at land surface; used only when Climate_Model is set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Land_Temperature_Filename": "Namawala_single_node_land_temperature_daily.bin"
}
|
| Load_Balance_Filename | string | NA | NA | UNINITIALIZED STRING | The path to the input file used when a static load balancing scheme is selected. The file must be in .json format. | {
"Load_Balance_Filename": "GitHub_426_LoadBalance.json"
}
|
| Local_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration to adjacent nodes by foot travel. The file must be in .bin format. | {
"Local_Migration_Filename": "Local_Migration.bin"
}
|
|
| Rainfall_Filename | string | NA | NA | rainfall.json | The path of the input file which defines rainfall data. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Rainfall_Filename": "Namawala_single_node_rainfall_daily.bin"
}
|
| Regional_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration by vehicle via road or rail network. If the node is not on a road or rail network, regional migration focuses on the closest hub city in the network. The file must be in .bin format. | {
"Regional_Migration_Filename": "Regional_Migration.bin"
}
|
|
| Relative_Humidity_Filename | string | NA | NA | rel_hum.json | The path of the input file which defines relative humidity data measured 2 meters above ground. Climate_Model must be set to CLIMATE_BY_DATA. The file must be in .bin format. | {
"Relative_Humidity_Filename": "Namawala_single_node_relative_humidity_daily.bin"
}
|
| Sea_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration by ship. Only used when Enable_Sea_Migration is set to true (1). The file must be in .bin format. | {
"Sea_Migration_Filename": "5x5_Households_Work_Migration.bin"
}
|
|
| Serialized_Population_Filenames | array of strings | NA | NA | NA | Array of filenames with serialized population data. The number of filenames must match the number of cores used for the simulation. The file must be in .dtk format. | {
"Serialized_Population_Filenames": [
"state-00010.dtk"
]
}
|
| Serialized_Population_Path | string | NA | NA | . | The root path for the serialized population files. | {
"Serialized_Population_Path": "../00_Generic_Version_1_save/output"
}
|
Migration¶
The following parameters determine aspects of population migration into and outside of a node, including daily commutes, seasonal migration, and one-way moves. Modes of transport includes travel by foot, automobile, sea, or air. Migration can also be configured to move all individuals in a family at the same time.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Air_Migration_Filename | string | NA | NA | The path to the input data file that defines patterns of migration by airplane. Enable_Air_Migration must be set to true (1). The file must be in .bin format. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration" : 1,
"Air_Migration_Filename": "../Global_1degree_air_migration.bin"
}
|
|
| Air_Migration_Roundtrip_Duration | float | 0 | 10000 | 1 | The average time spent (in days) at the destination node during a round-trip migration by airplane. Enable_Air_Migration must be set to true (1). | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration" : 1,
"Air_Migration_Roundtrip_Duration": 2
}
|
| Air_Migration_Roundtrip_Probability | float | 0 | 1 | 0.8 | The likelihood that an individual who flies to another node will return to the node of origin during the next migration. Enable_Air_Migration must be set to true (1). | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration" : 1,
"Air_Migration_Roundtrip_Probability": 0.9
}
|
| Enable_Air_Migration | boolean | 0 | 1 | 0 | Controls whether or not migration by air travel will occur. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Air_Migration": 1,
"Air_Migration_Filename": "../inputs/air_migration.bin"
}
|
| Enable_Family_Migration | boolean | 0 | 1 | 0 | Controls whether or not all members of a household can migrate together. All residents must be home before they can leave on the trip. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Enable_Migration": "FIXED_RATE_MIGRATION",
"Enable_Family_Migration": 1,
"Family_Migration_Filename": "../inputs/family_migration.bin"
}
|
| Enable_Local_Migration | boolean | 0 | 1 | 0 | Controls whether or not local migration (the diffusion of people in and out of nearby nodes by foot travel) occurs. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Local_Migration": 1,
"Local_Migration_Filename": "../inputs/local_migration.bin"
}
|
| Enable_Migration_Heterogeneity | boolean | 0 | 1 | 1 | Controls whether or not migration rate is heterogeneous among individuals. Set to true (1) to use a migration rate distribution in the demographics file (see NodeAttributes parameters); set to false (0) to use the same migration rate applied to all individuals. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Migration_Heterogeneity": 1
}
|
| Enable_Regional_Migration | boolean | 0 | 1 | 0 | Controls whether or not there is migration by road vehicle into and out of nodes in the road network. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Regional_Migration": 1,
"Regional_Migration_Filename": "../inputs/regional_migration.bin"
}
|
| Enable_Sea_Migration | boolean | 0 | 1 | 0 | Controls whether or not there is migration on ships into and out of coastal cities with seaports. Migration_Model must be set to FIXED_RATE_MIGRATION. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Sea_Migration": 1,
"Sea_Migration_Filename": "../inputs/sea_migration.bin"
}
|
| Family_Migration_Filename | string | NA | NA | The name of the binary file to use to configure family migration. Enable_Family_Migration must be set to true (1). The file must be in .bin format. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Enable_Family_Migration" 1,
"Family_Migration_Filename": "../inputs/family_migration.bin"
}
|
|
| Family_Migration_Roundtrip_Duration | float | 0 | 10000 | 1 | The number of days to complete the trip and return to the original node. Migration_Pattern must be set to SINGLE_ROUND_TRIPS. | {
"Migration_Model": "FIXED_RATE_MIGRATION",
"Migration_Pattern": "SINGLE_ROUND_TRIPS",
"Family_Migration_Roundtrip_Duration": 100
}
|
| Local_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration to adjacent nodes by foot travel. The file must be in .bin format. | {
"Local_Migration_Filename": "Local_Migration.bin"
}
|
|
| Local_Migration_Roundtrip_Duration | float | 0 | 10000 | 1 | The average time spent (in days) at the destination node during a round-trip migration by foot travel. Only used if Enable_Local_Migration is set to true (1). | {
"Local_Migration_Roundtrip_Duration": 1.0
}
|
| Local_Migration_Roundtrip_Probability | float | 0 | 1 | 0.95 | The likelihood that an individual who walks into a neighboring cell will return to the cell of origin during the next migration. Only used when Enable_Local_Migration is set to true (1). | {
"Local_Migration_Roundtrip_Probability": 1.0
}
|
| Migration_Model | enum | NA | NA | NO_MIGRATION | Model to use for migration. Possible values are:
|
{
"Migration_Model": "FIXED_RATE_MIGRATION",
"Local_Migration_Filename": "../inputs/local_migration.bin",
"Enable_Local_Migration": 1
}
|
| Migration_Pattern | enum | NA | NA | RANDOM_WALK_DIFFUSION | Describes the type of roundtrip used during migration. Migration_Model must be set to FIXED_RATE_MIGRATION. Possible values are:
|
{
"Migration_Model": "FIXED_RATE_MIGRATION",
"Migration_Pattern": "SINGLE_ROUND_TRIPS"
}
|
| Regional_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration by vehicle via road or rail network. If the node is not on a road or rail network, regional migration focuses on the closest hub city in the network. The file must be in .bin format. | {
"Regional_Migration_Filename": "Regional_Migration.bin"
}
|
|
| Regional_Migration_Roundtrip_Duration | float | 0 | 10000 | 1 | The average time spent (in days) at the destination node during a round-trip migration by road network. Enable_Regional_Migration must be set to true (1). | {
"Regional_Migration_Roundtrip_Duration": 1.0
}
|
| Regional_Migration_Roundtrip_Probability | float | 0 | 1 | 0.1 | The likelihood that an individual who travels by vehicle to another cell will return to the cell of origin during the next migration. Migration_Pattern must be set to SINGLE_ROUND_TRIPS. | {
"Regional_Migration_Roundtrip_Probability": 1.0
}
|
| Roundtrip_Waypoints | integer | 0 | 1000 | 10 | The maximum number of points reached during a trip before steps are retraced on the return trip home. Migration_Pattern must be set to WAYPOINTS_HOME. | {
"Roundtrip_Waypoints": 5
}
|
| Sea_Migration_Filename | string | NA | NA | The path of the input file which defines patterns of migration by ship. Only used when Enable_Sea_Migration is set to true (1). The file must be in .bin format. | {
"Sea_Migration_Filename": "5x5_Households_Work_Migration.bin"
}
|
|
| Sea_Migration_Roundtrip_Duration | float | 0 | 10000 | 1 | The average time spent at the destination node during a round-trip migration by ship. Used only when Enable_Sea_Migration is set to true (1). | {
"Sea_Migration_Roundtrip_Duration": 10000
}
|
| Sea_Migration_Roundtrip_Probability | float | 0 | 1 | 0.25 | The likelihood that an individual who travels by ship into a neighboring cell will return to the cell of origin during the next migration. Used only when Enable_Sea_Migration is set to true (1). | {
"Sea_Migration_Roundtrip_Probability": 0
}
|
| x_Air_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by air, as provided by the migration file. Enable_Air_Migration must be set to 1. | {
"x_Air_Migration": 1
}
|
| x_Family_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by families, as provided by the migration file. Enable_Family_Migration must be set to true (1). | {
"x_Family_Migration": 1
}
|
| x_Local_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for rate of migration by foot travel, as provided by the migration file. Enable_Local_Migration must be set to 1. | {
"x_Local_Migration": 1
}
|
| x_Regional_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by road vehicle, as provided by the migration file. Enable_Regional_Migration must be set to 1. | {
"x_Regional_Migration": 1
}
|
| x_Sea_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by sea, as provided by the migration file. Enable_Sea_Migration must be set to 1. | {
"x_Sea_Migration": 1
}
|
Mortality and survival¶
The following parameter determine mortality and survival characteristics of the disease being modeled and the population in general (non-disease mortality).
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| ART_CD4_at_Initiation_Saturating_Reduction_in_Mortality | float | 0 | 3.40E+38 | 350 | The duration from ART enrollment to on-ART HIV-caused death increases with CD4 at ART initiation up to a threshold determined by this parameter value. | {
"ART_CD4_at_Initiation_Saturating_Reduction_in_Mortality": 350
}
|
| Base_Mortality | float | 0 | 1000 | 0.001 | The base mortality of the infection before accounting for individual immune modification factors. Depending on the setting of Mortality_Time_Course, this is either the daily probability of the disease being fatal (DAILY_MORTALITY) or the probability of death at the end of the infection duration (MORTALITY_AFTER_INFECTIOUS). Enable_Vital_Dynamics must be set to true (1). | {
"Enable_Vital_Dynamics": 1,
"Mortality_Time_Course": "DAILY_MORTALITY",
"Base_Mortality": 0.01
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity | float | 0.1 | 10 | 1 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the heterogeneity (inverse shape) of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity": 0.5
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Scale | float | 1 | 3650 | 183 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the scale of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Scale": 618.3
}
|
| Death_Rate_Dependence | enum | NA | NA | NONDISEASE_MORTALITY_OFF | Determines how likely individuals are to die from natural, non-disease causes. Enable_Vital_Dynamics must be set to 1. Possible values are:
Properties, rates, and bin sizes can be set for non-disease mortality for each gender in the demographics file (see Complex distributions parameters). |
{
"Death_Rate_Dependence": "NONDISEASE_MORTALITY_OFF"
}
|
| HIV_Adult_Survival_Scale_Parameter_Intercept | float | 0.001 | 1000 | 21.182 | This parameter determines the intercept of the scale parameter, λ, for the Weibull distribution used to determine HIV survival time. Survival time with untreated HIV infection depends on the age of the individual at the time of infection, and is drawn from a Weibull distribution with shape parameter (see HIV_Adult_Survival_Shape_Parameter) and scale parameter, λ. The scale parameter is allowed to vary linearly with age as follows: λ = HIV_Adult_Survival_Scale_Parameter_Intercept + HIV_Adult_Survival_Scale_ Parameter_Slope * Age (in years). |
{
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Adult_Survival_Scale_Parameter_Slope | float | -1000 | 1000 | -0.2717 | This parameter determines the slope of the scale parameter, λ, for the Weibull distribution used to determine HIV survival time. Survival time with untreated HIV infection depends on the age of the individual at the time of infection, and is drawn from a Weibull distribution with shape parameter (see HIV_Adult_Survival_Shape_Parameter) and scale parameter, λ. The scale parameter is allowed to vary linearly with age as follows: λ = HIV_Adult_Survival_Scale_Parameter_Intercept + HIV_Adult_Survival_Scale_ Parameter_Slope * Age (in years). Because survival time with HIV becomes shorter with increasing age, HIV_Adult_Survival_Scale_ Parameter_Slope should be set to a negative number. |
{
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Adult_Survival_Shape_Parameter | float | 0.001 | 1000 | 2 | This parameter determines the shape of the Weibull distribution used to determine age-dependent survival time for individuals infected with HIV. | {
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Age_Max_for_Adult_Age_Dependent_Survival | float | 0 | 75 | 70 | Survival time with untreated HIV infection depends on the age of the individual at the time of infection, and is drawn from a Weibull distribution with shape parameter and scale parameter, λ. See HIV_Adult_Survival_Scale_Parameter_Intercept, HIV_Adult_Survival_Scale_ Parameter_Slope, and HIV_Adult_Survival_Shape_Parameter. Although the scale parameter for survival time declines with age, it cannot become negative. To avoid negative survival times at older ages, this parameter, HIV_Age_Max_for_Adult_Age_Dependent_Survival, determines the age beyond which HIV survival is no longer affected by further aging. |
{
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Age_Max_for_Child_Survival_Function | float | 0 | 75 | 15 | The maximum age at which an individual’s survival will be fit to the child survival function. If the value of this parameter falls between zero and the age of sexual debut, model results are not sensitive to this parameter as there is no mechanism for children to become infected between infancy and sexual debut. | {
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0,
"HIV_Age_Max_for_Child_Survival_Function": 15.0
}
|
| HIV_Child_Survival_Rapid_Progressor_Fraction | float | 0 | 1 | 0.57 | The proportion of HIV-infected children who are rapid HIV progressors. | {
"HIV_Child_Survival_Rapid_Progressor_Fraction": 0.57,
"HIV_Child_Survival_Rapid_Progressor_Rate": 1.52
}
|
| HIV_Child_Survival_Rapid_Progressor_Rate | float | 0 | 1000 | 1.52 | The exponential decay rate, in years, describing the distribution of HIV survival for children who are rapid progressors. | {
"HIV_Child_Survival_Rapid_Progressor_Fraction": 0.57,
"HIV_Child_Survival_Rapid_Progressor_Rate": 1.52
}
|
| HIV_Child_Survival_Slow_Progressor_Scale | float | 0.001 | 1000 | 16 | The Weibull scale parameter describing the distribution of HIV survival for children who are slower progressors. | {
"HIV_Child_Survival_Slow_Progressor_Scale": 16.0,
"HIV_Child_Survival_Slow_Progressor_Shape": 2.7
}
|
| HIV_Child_Survival_Slow_Progressor_Shape | float | 0.001 | 1000 | 2.7 | The Weibull shape parameter describing the distribution of HIV survival for children who are slower progressors. | {
"HIV_Child_Survival_Slow_Progressor_Scale": 16.0,
"HIV_Child_Survival_Slow_Progressor_Shape": 2.7
}
|
| Mortality_Blocking_Immunity_Decay_Rate | float | 0 | 1000 | 0.001 | The rate at which mortality-blocking immunity decays after the mortality-blocking immunity offset period. Enable_Immune_Decay must be set to 1. | {
"Mortality_Blocking_Immunity_Decay_Rate": 0.1
}
|
| Mortality_Time_Course | enum | NA | NA | DAILY_MORTALITY | The method used to calculate disease deaths. Possible values are:
|
{
"Mortality_Time_Course": "MORTALITY_AFTER_INFECTIOUS"
}
|
| x_Other_Mortality | float | 0 | 3.40E+38 | 1 | Scale factor for mortality from causes other than the disease being simulated, as provided by the demographics file (see Complex distributions parameters). Enable_Vital_Dynamics must be set to 1. | {
"x_Other_Mortality": 1
}
|
Output settings¶
The following parameters configure whether or not output reports are created for the simulation, such as reports detailing spatial or demographic data at each time step. By default, the Inset chart output report is always created.
The following figures are examples for the parameter Report_HIV_Period.
When Report_HIV_Period is set to a value that is less than the Simulation_Timestep, a record will be written during the next time step after the reported period. More than one period may occur before the next time step.
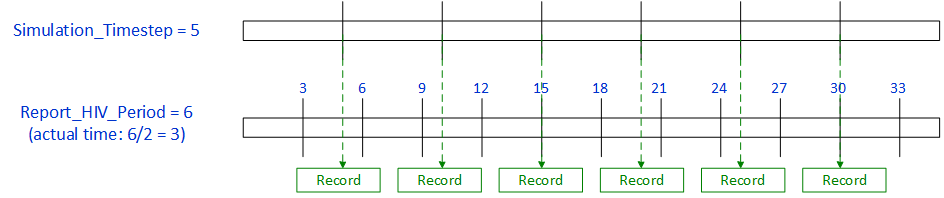
Figure 1: Report_HIV_Period < **Simulation_Timestep
When Report_HIV_Period is greater than Simulation_Timestep, a record will be written during the next time step after the period occurs. This means that a record may not be written at all time steps.
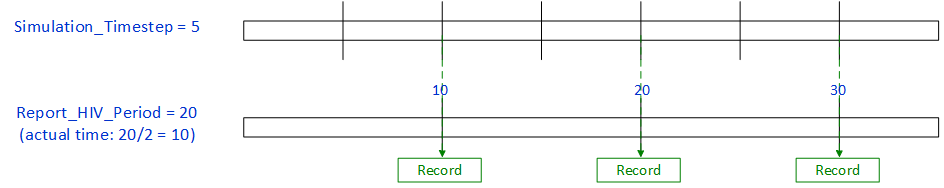
Figure 2: Report_HIV_Period > **Simulation_Timestep
| Parameter | Data type | Minimum | Maximum | Default | Description | Example | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Custom_Reports_Filename | string | NA | NA | UNINITIALIZED STRING | The name of the file containing custom report configuration parameters. Omitting this parameter or setting it to RunAllCustomReports will load all reporters found that are valid for the given simulation type. The file must be in JSON format. | {
"Custom_Reports_Filename": "custom_reports.json"
}
|
|||||||||||||||
| Enable_Default_Reporting | boolean | 0 | 1 | 1 | Controls whether or not the default InsetChart.json report is created. | {
"Enable_Default_Reporting": 1
}
|
|||||||||||||||
| Enable_Demographics_Reporting | boolean | 0 | 1 | 1 | Controls whether or not demographic summary data and age-binned reports are outputted to file. | {
"Enable_Demographics_Reporting": 1
}
|
|||||||||||||||
| Enable_Property_Output | boolean | 0 | 1 | 0 | Controls whether or not to create property output reports, which detail groups as defined in IndividualProperties in the demographics file (see NodeProperties and IndividualProperties parameters). When there is more than one property type, the report will display the channel information for all combinations of the property type groups. | {
"Enable_Property_Output": 1
}
|
|||||||||||||||
| Enable_Spatial_Output | boolean | 0 | 1 | 0 | Controls whether or not spatial output reports are created. If set to true (1), spatial output reports include all channels listed in the parameter Spatial_Output_Channels. Note Spatial output files require significant processing time and disk space. |
{
"Enable_Spatial_Output": 1,
"Spatial_Output_Channels": [
"Prevalence",
"New_Infections"
]
}
|
|||||||||||||||
| Report_Coital_Acts | boolean | 0 | 1 | 0 | Set to true (1) to enable or to false (0) to disable the RelationshipConsummatedReport.csv output report. | {
"Report_Coital_Acts": 1
}
|
|||||||||||||||
| Report_Event_Recorder | boolean | 0 | 1 | 0 | Set to true (1) to enable or to false (0) to disable the ReportEventRecorder.csv output report. | {
"Report_Event_Recorder": 1,
"Report_Event_Recorder_Events": [
"VaccinatedA",
"VaccineExpiredA",
"VaccinatedB",
"VaccineExpiredB"
],
"Report_Event_Recorder_Ignore_Events_In_List": 0
}
|
|||||||||||||||
| Report_Event_Recorder_Events | array | NA | NA | The list of events to include or exclude in the ReportEventRecorder.csv output report, based on how Report_Event_Recorder_Ignore_Events_In_List is set. | {
"Report_Event_Recorder": 1,
"Report_Event_Recorder_Events": [
"VaccinatedA",
"VaccineExpiredA",
"VaccinatedB",
"VaccineExpiredB"
],
"Report_Event_Recorder_Ignore_Events_In_List": 0
}
|
||||||||||||||||
| Report_Event_Recorder_Ignore_Events_In_List | boolean | 0 | 1 | 0 | If set to false (0), only the events listed in the Report_Event_Recorder_Events array will be included in the ReportEventRecorder.csv output report. If set to true (1), only the events listed in the array will be excluded, and all other events will be included. If you want to return all events from the simulation, leave the events array empty.
|
{
"Report_Event_Recorder": 1,
"Report_Event_Recorder_Events": [
"VaccinatedA",
"VaccineExpiredA",
"VaccinatedB",
"VaccineExpiredB"
],
"Report_Event_Recorder_Ignore_Events_In_List": 0
}
|
|||||||||||||||
| Report_Event_Recorder_Individual_Properties | array of strings | NA | NA | [] | Specifies an array of events that will be excluded from the property output report; all events NOT listed in the array will be included in the report. To report all events from the simulation, leave the events array empty. | This example demonstrates reporting all individual property events: {
"Report_Event_Recorder_Individual_Properties": []
}
The following example demonstrates the syntax for excluding particular properties from the report: {
"Report_Event_Recorder_Individual_Properties": [
"Accessibility",
"Risk"
]
}
|
|||||||||||||||
| Report_HIV_ART | boolean | 0 | 1 | 0 | Set to true (1) to enable or to false (0) to disable the ReportHIVART.csv output report. | {
"Report_HIV_ART": 0
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender | boolean | 0 | 1 | 0 | Set to true (1) to enable or to false (0) to disable the ReportHIVByAgeAndGener.csv output report. | {
"Report_HIV_ByAgeAndGender": 1
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Add_Relationships | boolean | 0 | 1 | 0 | Sets whether or not the ReportHIVByAgeAndGender.csv output file will contain data by relationship type on population currently in a relationship and ever in a relationship. A sum of those in two or more partnerships and a sum of the lifetime number of relationships in each bin will be included. | {
"Report_HIV_ByAgeAndGender_Add_Relationships": 1
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Add_Transmitters | boolean | 0 | 1 | 0 | When Set to to true (1), the “transmitters” column is included in the output report. For a given row, “Transmitters” indicates how many people that have transmitted the disease meet the specifications of that row. | {
"Report_HIV_ByAgeAndGender_Add_Transmitters": 1
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data | array of floats | -3.40282e+38 | 3.40282e+38 | 1 | This array of floats allows the user to define the age bins used to stratify the report by age. Each value defines the minimum value of that bin, while the next value defines the maximum value of the bin. The maximum number of age bins is 100. For example, if you had: “Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data” : [ 0, 10, 20, 50, 100 ] The report would have the following age bins: From 0 up to (but not including) 10, from 10 up to (but not including) 20, from 20 up to (but not including) 50, from 50 up to (but not including) 100, and 100 and over. If no values are specified in the array, then the output report will have no age binning. |
{
"Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data" : [
0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93, 94, 95, 96, 97, 98, 99
]
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_Circumcision_Data | boolean | 0 | 1 | 0 | When set to 1, the ReportHIVByAgeAndGender.csv output report will have separate rows for those who do or do not have the MaleCircumcision intervention and an additional column with 0 and 1 indicating whether the row corresponds to those who are or are not circumcised. Setting this to 1 doubles the number of rows in ReportHIVByAgeAndGender.csv. | {
"Report_HIV_ByAgeAndGender_Collect_Circumcision_Data": 0
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_Gender_Data | boolean | 0 | 1 | 0 | Controls whether or not the report is stratified by gender; to enable gender stratification, set to true (1). | {
"Report_HIV_ByAgeAndGender_Collect_Gender_Data": 1
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_HIV_Data | boolean | 0 | 1 | 0 | When set to 1, the ReportHIVByAgeAndGender.csv output report will have separate rows for those who do or do not have the ART intervention and an additional column with 0 and 1 indicating whether the row corresponds to those who are or are not on ART. Setting this to 1 doubles the number of rows in ReportHIVByAgeAndGender.csv. | {
"Report_HIV_ByAgeAndGender_Collect_HIV_Data": 1
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_Intervention_Data | array of strings | NA | NA | NA | Stratifies the population by adding a column of 0s and 1s depending on whether or not the person has the indicated intervention. This only works for interventions that remain with a person for a period of time, such as ART, VMMC, vaccine/PrEP, or a delay state in the cascade of care. You can name the intervention by adding a parameter Intervention_Name in the campaign file, and then give this parameter the same Intervention_Name. This allows you to have multiple types of vaccines, VMMCs, etc., but to only stratify on the type you want. | {
"Report_HIV_ByAgeAndGender_Collect_Intervention_Data": [
"ART_Intervention",
"PrEP_Intervention"
]
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Collect_IP_Data | boolean | 0 | 1 | 0 | When set to 1, the ReportHIVByAgeAndGender.csvoutput report will have separate rows for those with different IndividualProperties values and an additional column for each property indicating which row corresponds to which value of that property. Setting this to 1 typically increases by severalfold the number of rows in ReportHIVByAgeAndGender.csv. | {
"Report_HIV_ByAgeAndGender_Collect_IP_Data": 0
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Event_Counter_List | array of strings | NA | NA | NA | A list of columns to add to the ReportHIVByAgeAndGender.csv output files counting the number of times an intervention with the corresponding Distributed_Event_Trigger has been distributed. Note that the interventions will be specified in the campaign file. | {
"Report_HIV_ByAgeAndGender_Event_Counter_List": [
"Reduced_Acquire_Lowest",
"Reduced_Acquire_Medium",
"Reduced_Acquire_Low",
"Reduced_Acquire_Highest_Not_Duplicated"
]
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Start_Year | float | 1900 | 2200 | 1900 | The beginning calendar year that will be collected by the ReportHIVByAgeAndGender.csv report. | {
"Report_HIV_ByAgeAndGender_Start_Year": 1962,
"Report_HIV_ByAgeAndGender_Stop_Year": 1978
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Stop_Year | float | 1900 | 2200 | 2200 | The ending calendar year that will be collected by the HIVByAgeAndGender.csv report. | {
"Report_HIV_ByAgeAndGender_Start_Year": 1962,
"Report_HIV_ByAgeAndGender_Stop_Year": 1978
}
|
|||||||||||||||
| Report_HIV_ByAgeAndGender_Stratify_Infected_By_CD4 | boolean | 0 | 1 | 0 | When set to 1, the ReportHIVByAgeAndGender.csv output file will separate the count of the number of infected individuals by CD4 stratum. When set to 0, the number of infected individuals are aggregated into one column regardless of CD4 count. | {
"Report_HIV_ByAgeAndGender_Stratify_Infected_By_CD4": 0
}
|
|||||||||||||||
| Report_HIV_Event_Channels_List | array of strings | NA | NA | NA | This is the list of events included in the InsetChart report. If events are specified with this parameter, the InsetChart will include a channel for each event listed. If no events are listed, a “Number of Events” channel will display the total number of all events that occurred during the simulation. | {
"Report_HIV_Event_Channels_List": [
"NewInfectionEvent",
"HIVNeedsHIVTest",
"HIVPositiveHIVTest",
"HIVNegativeHIVTest",
"HIVDiagnosedEligibleForART",
"HIVSymptomatic",
"DiseaseDeaths",
"NonDiseaseDeaths",
"Births"
],
}
|
|||||||||||||||
| Report_HIV_Infection | boolean | 0 | 1 | 0 | Enables or disables the ReportHIVInfection.csv output report. | {
"Report_HIV_Infection": 0
}
|
|||||||||||||||
| Report_HIV_Infection_Start_Year | float | 1900 | 2200 | 1900 | The beginning calendar year that will be collected by the ReportHIVInfection.csv output report. Report_HIV_Infection must be set to 1. | {
"Report_HIV_Infection_Start_Year": 1900
}
|
|||||||||||||||
| Report_HIV_Infection_Stop_Year | float | 1900 | 2200 | 2200 | The ending calendar year that will be collected by the ReportHIVInfection.csv output report. Report_HIV_Infection must be set to 1. | {
"Report_HIV_Infection_Stop_Year": 2100
}
|
|||||||||||||||
| Report_HIV_Mortality | boolean | 0 | 1 | 0 | Enables or disables the HIVMortality.csv (disease and non-disease deaths) output report. | {
"Report_HIV_Mortality": 0
}
|
|||||||||||||||
| Report_HIV_Period | float | 30 | 36500 | 730 | The number of days between records in the HIV_By_Age_And_Gender output report. Output data will only be recorded during a time step, so if Report_HIV_Period is set to a value less than the value set for Simulation_Timestep, more than one period may occur before the next time step. When Report_HIV_Period is greater than the value set for Simulation_Timestep, a record may not be written during each time step. Note that the number of days between records is half the time specified by this parameter. For example, if Report_HIV_Period is set to 40, the actual time between records is 20 days. For best results, use integers for this value. | {
"Report_HIV_Period": 365
}
|
|||||||||||||||
| Report_Relationship_End | boolean | 0 | 1 | 0 | Enables or disables the RelationshipEnd.csv output report. For HIV simulations, there will be no additional columns. | {
"Report_Relationship_End": 0
}
|
|||||||||||||||
| Report_Relationship_Start | boolean | 0 | 1 | 0 | Enables or disables the RelationshipStart.csv output report. For HIV simulations, there will be some additional columns. | {
"Report_Relationship_Start": 0
}
|
|||||||||||||||
| Report_Transmission | boolean | 0 | 1 | 0 | Enables or disables the TransmissionReport.csv output report. For HIV simulations, there will be some additional columns. | {
"Report_Transmission": 0
}
|
|||||||||||||||
| Spatial_Output_Channels | array of strings | NA | NA | [] | An array of channel names for spatial output by node and time step. The data from each channel will be written to a separate binary file. Enable_Spatial_Output must be set to true (1). Possible values are:
|
{
"Spatial_Output_Channels": [
"Prevalence",
"New_Infections"
]
}
|
Population dynamics¶
The following parameters determine characteristics related to population dynamics, such as age distribution, births, deaths, and gender. The values set here generally interact closely with values in the demographics file.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Age_Initialization_Distribution_Type | enum | NA | NA | DISTRIBUTION_OFF | The method for initializing the age distribution in the simulated population. Possible values are:
|
{
"Age_Initialization_Distribution_Type": "DISTRIBUTION_SIMPLE"
}
|
| Base_Population_Scale_Factor | float | 0 | 3.40E+38 | 1 | The scale factor for InitialPopulation in the demographics file (see NodeAttributes parameters). If Population_Scale_Type is set to FIXED_SCALING, the initial simulation population is uniformly scaled over the entire area to adjust for historical or future population density. | {
"Base_Population_Scale_Factor": 0.0001
}
|
| Birth_Rate_Boxcar_Forcing_Amplitude | float | 0 | 3.40E+38 | 0 | Fractional increase in birth rate during high birth season when Birth_Rate_Time_Dependence is set to ANNUAL_BOXCAR_FUNCTION. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "ANNUAL_BOXCAR_FUNCTION",
"Birth_Rate_Boxcar_Forcing_Amplitude": 0.1
}
|
| Birth_Rate_Boxcar_Forcing_End_Time | float | 0 | 365 | 0 | Day of the year when the high birth rate season ends when Birth_Rate_Time_Dependence is set to ANNUAL_BOXCAR_FUNCTION. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "ANNUAL_BOXCAR_FUNCTION",
"Birth_Rate_Boxcar_Forcing_End_Time": 220
}
|
| Birth_Rate_Boxcar_Forcing_Start_Time | float | 0 | 365 | 0 | Day of the year when the high birth rate season begins when Birth_Rate_Time_Dependence is set to ANNUAL_BOXCAR_FUNCTION. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "ANNUAL_BOXCAR_FUNCTION",
"Birth_Rate_Boxcar_Forcing_Start_Time": 130
}
|
| Birth_Rate_Dependence | enum | NA | NA | FIXED_BIRTH_RATE | The method used to modify the value set in BirthRate in the demographics file (see NodeAttributes parameters). Possible values are:
|
{
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Dependence": "POPULATION_DEP_RATE"
}
|
| Birth_Rate_Sinusoidal_Forcing_Amplitude | float | 0 | 1 | 0 | The amplitude of sinusoidal variations in birth rate when Birth_Rate_Time_Dependence is set to SINUSOIDAL_FUNCTION_OF_TIME. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "SINUSOIDAL_FUNCTION_OF_TIME",
"Birth_Rate_Sinusoidal_Forcing_Amplitude": 0.1
}
|
| Birth_Rate_Sinusoidal_Forcing_Phase | float | 0 | 365 | 0 | The phase of sinusoidal variations in birth rate. Birth_Rate_Time_Dependence must be set to SINUSOIDAL_FUNCTION_OF_TIME. | {
"Birth_Rate_Sinusoidal_Forcing_Phase": 20
}
|
| Birth_Rate_Time_Dependence | enum | NA | NA | NONE | A scale factor that allows the birth rate to be altered by time or season. Enable_Birth must be set to true (1). Possible values are:
|
{
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "ANNUAL_BOXCAR_FUNCTION"
}
|
| Death_Rate_Dependence | enum | NA | NA | NONDISEASE_MORTALITY_OFF | Determines how likely individuals are to die from natural, non-disease causes. Enable_Vital_Dynamics must be set to 1. Possible values are:
Properties, rates, and bin sizes can be set for non-disease mortality for each gender in the demographics file (see Complex distributions parameters). |
{
"Death_Rate_Dependence": "NONDISEASE_MORTALITY_OFF"
}
|
| Default_Geography_Initial_Node_Population | integer | 0 | 1000000 | 1000 | When using the built-in demographics for default geography, the initial number of individuals in each node. Note that the built-in demographics feature does not represent a real geographical location and is mostly used for testing. Enable_Demographics_Builtin must be set to true (1). | {
"Enable_Demographics_Builtin": 1,
"Default_Geography_Initial_Node_Population": 1000,
"Default_Geography_Torus_Size": 3
}
|
| Demographics_Filenames | array of strings | NA | NA | An array of the paths to demographics files containing information on the identity and demographics of the region to simulate. The files must be in .json format. | {
"Demographics_Filenames": [
"Namawala_single_node_demographics.json",
"Namawala_demographics_overlay.json"
]
}
|
|
| Enable_Aging | boolean | 0 | 1 | 1 | Controls whether or not individuals in a population age during the simulation. Enable_Vital_Dynamics must be set to true (1). | {
"Enable_Vital_Dynamics": 1,
"Enable_Aging": 1
}
|
| Enable_Birth | boolean | 0 | 1 | 1 | Controls whether or not individuals will be added to the simulation by birth. Enable_Vital_Dynamics must be set to true (1). If you want new individuals to have the same intervention coverage as existing individuals, you must add a BirthTriggeredIV to the campaign file. | {
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1
}
|
| Enable_Demographics_Birth | boolean | 0 | 1 | 0 | Controls whether or not newborns have identical or heterogeneous characteristics. Set to false (0) to give all newborns identical characteristics; set to true (1) to allow for heterogeneity in traits such as sickle-cell status. Enable_Birth must be set to true (1). | {
"Enable_Birth": 1,
"Enable_Demographics_Birth": 1
}
|
| Enable_Demographics_Builtin | boolean | 0 | 1 | 0 | Controls whether or not built-in demographics for default geography will be used. Note that the built-in demographics feature does not represent a real geographical location and is mostly used for testing. Set to true (1) to define the initial population and number of nodes using Default_Geography_Initial_Node_Population and Default_Geography_Torus_Size. Set to false (0) to use demographics input files defined in Demographics_Filenames. | {
"Enable_Demographics_Builtin": 1,
"Default_Geography_Initial_Node_Population": 1000,
"Default_Geography_Torus_Size": 3
}
|
| Enable_Demographics_Gender | boolean | 0 | 1 | 1 | Controls whether or not gender ratios are drawn from a Gaussian or 50/50 draw. Set to true (1) to create gender ratios drawn from a male/female ratio that is randomly smeared by a Gaussian of width 1%; set to false (0) to assign a gender ratio based on a 50/50 draw. | {
"Enable_Demographics_Gender": 1
}
|
| Enable_Demographics_Other | boolean | 0 | 1 | 0 | Controls whether or not other demographic factors are included in the simulation, such as the fraction of individuals above poverty, urban/rural characteristics, heterogeneous initial immunity, or risk. These factors are set in the demographics file. | {
"Enable_Demographics_Other": 1
}
|
| Enable_Disease_Mortality | boolean | 0 | 1 | 1 | Controls whether or not individuals die due to disease. | {
"Enable_Disease_Mortality": 1
}
|
| Enable_Vital_Dynamics | boolean | 0 | 1 | 1 | Controls whether or not births and deaths occur in the simulation. Births and deaths must be individually enabled and set. | {
"Enable_Vital_Dynamics":1,
"Enable_Birth": 1,
"Death_Rate_Dependence": "NONDISEASE_MORTALITY_OFF",
"Base_Mortality": 0.002
}
|
| Minimum_Adult_Age_Years | float | 0 | 3.40E+38 | 15 | The age, in years, after which an individual is considered an adult. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP. | {
"Minimum_Adult_Age_Years": 17
}
|
| Population_Density_Infectivity_Correction | enum | NA | NA | CONSTANT_INFECTIVITY | Correction to alter infectivity by population density set in the Population_Density_C50 parameter. Measured in people per square kilometer. Possible values are:
Note Sparsely populated areas have a lower infectivity, while densely populated areas have a higher infectivity, which rises to saturate at the Base_Infectivity value. |
{
"Population_Density_Infectivity_Correction": "SATURATING_FUNCTION_OF_DENSITY"
}
|
| Population_Scale_Type | enum | NA | NA | USE_INPUT_FILE | The method to use for scaling the initial population specified in the demographics input file. Possible values are:
|
{
"Population_Scale_Type": "FIXED_SCALING"
}
|
| Report_HIV_ByAgeAndGender_Add_Transmitters | boolean | 0 | 1 | 0 | When Set to to true (1), the “transmitters” column is included in the output report. For a given row, “Transmitters” indicates how many people that have transmitted the disease meet the specifications of that row. | {
"Report_HIV_ByAgeAndGender_Add_Transmitters": 1
}
|
| Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data | array of floats | -3.40282e+38 | 3.40282e+38 | 1 | This array of floats allows the user to define the age bins used to stratify the report by age. Each value defines the minimum value of that bin, while the next value defines the maximum value of the bin. The maximum number of age bins is 100. For example, if you had: “Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data” : [ 0, 10, 20, 50, 100 ] The report would have the following age bins: From 0 up to (but not including) 10, from 10 up to (but not including) 20, from 20 up to (but not including) 50, from 50 up to (but not including) 100, and 100 and over. If no values are specified in the array, then the output report will have no age binning. |
{
"Report_HIV_ByAgeAndGender_Collect_Age_Bins_Data" : [
0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93, 94, 95, 96, 97, 98, 99
]
}
|
| Report_HIV_ByAgeAndGender_Collect_Gender_Data | boolean | 0 | 1 | 0 | Controls whether or not the report is stratified by gender; to enable gender stratification, set to true (1). | {
"Report_HIV_ByAgeAndGender_Collect_Gender_Data": 1
}
|
| x_Birth | float | 0 | 3.40E+38 | 1 | Scale factor for birth rate, as provided by the demographics file (see NodeAttributes parameters). Enable_Birth must be set to 1. | {
"x_Birth": 1
}
|
Relationships and pair formation¶
The following parameters determine how individuals form relationships, such as sexual behavior, relationship duration, age gaps, and any gender differences in those characteristics.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Coital_Dilution_Factor_2_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly two current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_2_Partners" 0.5
}
|
| Coital_Dilution_Factor_3_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly three current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_3_Partners": 0.33
}
|
| Coital_Dilution_Factor_4_Plus_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly three current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_4_Partners": 0.45
}
|
| Enable_Coital_Dilution | boolean | 0 | 1 | 1 | Controls whether or not coital dilution will occur. | {
"Enable_Coital_Dilution": 1
}
|
| Min_Days_Between_Adding_Relationships | float | 0 | 365 | 60 | The minimum number of days between adding two consecutive relationships as a means to control concurrency. The constraint does not apply if an individual breaks an existing relationship. | {
"Min_Days_Between_Adding_Relationships": 1
}
|
| PFA_Burnin_Duration_In_Days | float | 1 | 3.40E+38 | 365000 | The number of days to continue tuning the pair formation rates. After this duration, the rates will remain at the last rate value calculated. | {
"PFA_Burnin_Duration_In_Days": 5475
}
|
| PFA_Cum_Prob_Selection_Threshold | float | 0 | 1 | 0.2 | This parameter serves to minimize the extent to which relationships with unlikely age gaps are formed. These unlikely relationships could be generated due to lack of diversity in the PFA. Within the algorithm, males pick from amongst the available females by age bin, weighted by the conditional of the joint_probability matrix given the male age bin. If the sum of the probabilities in the female age bins that are not empty is below this threshold, the male will wait till the next update. Setting this parameter to a value near 1 dramatically increases the delay that individuals will experience in seeking relationships. Setting this parameter to 0 disables the feature. | {
"PFA_Cum_Prob_Selection_Threshold": 0.4
}
|
| Report_HIV_ByAgeAndGender_Add_Relationships | boolean | 0 | 1 | 0 | Sets whether or not the ReportHIVByAgeAndGender.csv output file will contain data by relationship type on population currently in a relationship and ever in a relationship. A sum of those in two or more partnerships and a sum of the lifetime number of relationships in each bin will be included. | {
"Report_HIV_ByAgeAndGender_Add_Relationships": 1
}
|
| Sexual_Debut_Age_Female_Weibull_Heterogeneity | float | 0 | 50 | 20 | The inverse shape of the Weibull distribution for female debut age. | {
"Sexual_Debut_Age_Female_Weibull_Heterogeneity": 0.05
}
|
| Sexual_Debut_Age_Female_Weibull_Scale | float | 0 | 50 | 16 | The scale term of the Weibull distribution for female debut age. | {
"Sexual_Debut_Age_Female_Weibull_Scale": 15.919654846191
}
|
| Sexual_Debut_Age_Male_Weibull_Heterogeneity | float | 0 | 50 | 20 | The inverse shape of the Weibull distribution for male debut age. | {
"Sexual_Debut_Age_Male_Weibull_Heterogeneity": 0.05
}
|
| Sexual_Debut_Age_Male_Weibull_Scale | float | 0 | 50 | 16 | The scale term of the Weibull distribution for male debut age. | {
"Sexual_Debut_Age_Male_Weibull_Scale": 16.946729660034
}
|
| Sexual_Debut_Age_Min | float | 0 | 3.40E+38 | 13 | The minimum age at which individuals become eligible to form sexual relationships. | {
"Sexual_Debut_Age_Min": 13
}
|
Sampling¶
The following parameters determine how a population is sampled in the simulation. While you may want every agent (individual object) to represent a single person, you can often optimize CPU time with without degrading the accuracy of the simulation but having an agent represent multiple people. The sampling rate may be adapted to have a higher or lower sampling rate for particular regions or age groups.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Base_Individual_Sample_Rate | float | 0 | 1 | 1 | The base rate of sampling for individuals, equal to the fraction of individuals in each node being sampled. Reducing the sampling rate will reduce the time needed to run simulations. Individual_Sampling_Type must be set to FIXED_SAMPLING or ADAPTED_SAMPLING_BY_IMMUNE_STATE. | {
"Base_Individual_Sample_Rate": 0.01
}
|
| Immune_Threshold_For_Downsampling | float | 0 | 1 | 0 | Threshold on acquisition immunity at which to apply immunity dependent downsampling. Individual_Sampling_Type must set to ADAPTED_SAMPLING_BY_IMMUNE_STATE. | {
"Individual_Sampling_Type": "ADAPTED_SAMPLING_BY_IMMUNE_STATE",
"Immune_Threshold_For_Downsampling": 0.5
}
|
| Individual_Sampling_Type | enum | NA | NA | TRACK_ALL | The type of individual human sampling. Possible values are:
|
The following example shows how to sampling immune individuals at a lower rate. {
"Individual_Sampling_Type": "ADAPTED_SAMPLING_BY_IMMUNE_STATE",
"Immune_Threshold_For_Downsampling": 0.5,
"Relative_Sample_Rate_Immune": 0.1
}
The following example shows how to sampling older individuals at a lower rate. {
"Individual_Sampling_Type": "ADAPTED_SAMPLING_BY_AGE_GROUP",
"Sample_Rate_0_18mo": 1,
"Sample_Rate_10_14": 0.5,
"Sample_Rate_15_19": 0.3,
"Sample_Rate_18mo_4yr": 1,
"Sample_Rate_20_Plus": 0.2,
"Sample_Rate_5_9": 1,
"Sample_Rate_Birth": 1
}
|
| Max_Node_Population_Samples | float | 1 | 3.40E+38 | 30 | The maximum number of individuals to track in a node. When the population exceeds this number, the sampling rate will drop according to the value set in Individual_Sampling_Type. | {
"Individual_Sampling_Type": "ADAPTED_SAMPLING_BY_POPULATION_SIZE",
"Max_Node_Population_Samples": 100000
}
|
| Relative_Sample_Rate_Immune | float | 0.001 | 1 | 0.1 | The relative sampling rate for people who have acquired immunity through recovery or vaccination. | {
"Relative_Sample_Rate_Immune": 0.1
}
|
| Sample_Rate_0_18mo | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for infants age 0 to 18 months. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_0_18mo": 1
}
|
| Sample_Rate_10_14 | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for children age 10 to 14 years. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_10_14": 1
}
|
| Sample_Rate_15_19 | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for adolescents age 15 to 19 years. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_15_19": 1
}
|
| Sample_Rate_18mo_4yr | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for children age 18 months to 4 years. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_18mo_4yr": 1
}
|
| Sample_Rate_20_plus | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for adults age 20 and older. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_20_plus": 1
}
|
| Sample_Rate_5_9 | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for children age 5 to 9 years. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_5_9": 1
}
|
| Sample_Rate_Birth | float | 0 | 1000 | 1 | For age-adapted sampling, the relative sampling rate for births. Individual_Sampling_Type must be set to ADAPTED_SAMPLING_BY_AGE_GROUP or ADAPTED_SAMPLING_BY_AGE_GROUP_AND_POP_SIZE. | {
"Sample_Rate_Birth": 1
}
|
Scalars and multipliers¶
The following parameters scale or multiply values set in other areas of the configuration file or input data files. This can be especially useful for understanding the sensitivities of disease dynamics to input data without requiring modifications to those base values. For example, one might set x_Birth to a value less than 1 to simulate a lower future birth rate due to increased economic prosperity and available medical technology.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Acute_Stage_Infectivity_Multiplier | float | 1 | 100 | 26 | The multiplier acting on Base_Infectivity to determine the per-act transmission probability of an HIV+ individual during the acute stage. | {
"Acute_Stage_Infectivity_Multiplier": 3
}
|
| AIDS_Stage_Infectivity_Multiplier | float | 1 | 100 | 10 | The multiplier acting on Base_Infectivity to determine the per-act transmission probability of an HIV+ individual during the AIDS stage. | {
"AIDS_Stage_Infectivity_Multiplier": 8
}
|
| ART_Viral_Suppression_Multiplier | float | 0 | 1 | 0.08 | Multiplier acting on Base_Infectivity to determine the per-act transmission probability of a virally suppressed HIV+ individual. | {
"ART_Viral_Suppression_Multiplier": 0.3
}
|
| Base_Population_Scale_Factor | float | 0 | 3.40E+38 | 1 | The scale factor for InitialPopulation in the demographics file (see NodeAttributes parameters). If Population_Scale_Type is set to FIXED_SCALING, the initial simulation population is uniformly scaled over the entire area to adjust for historical or future population density. | {
"Base_Population_Scale_Factor": 0.0001
}
|
| Birth_Rate_Time_Dependence | enum | NA | NA | NONE | A scale factor that allows the birth rate to be altered by time or season. Enable_Birth must be set to true (1). Possible values are:
|
{
"Enable_Vital_Dynamics": 1,
"Enable_Birth": 1,
"Birth_Rate_Time_Dependence": "ANNUAL_BOXCAR_FUNCTION"
}
|
| CD4_At_Death_LogLogistic_Scale | float | 1 | 1000 | 31.63 | The scale parameter of a Weibull distribution that represents the at-death CD4 cell count. | {
"CD4_At_Death_LogLogistic_Scale": 2.96
}
|
| CD4_Post_Infection_Weibull_Scale | float | 1 | 1000 | 560.432 | The scale parameter of a Weibull distribution that represents the post-acute-infection CD4 cell count. | {
"CD4_Post_Infection_Weibull_Scale": 550
}
|
| Coital_Dilution_Factor_2_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly two current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_2_Partners" 0.5
}
|
| Coital_Dilution_Factor_3_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly three current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_3_Partners": 0.33
}
|
| Coital_Dilution_Factor_4_Plus_Partners | float | 1.19E-0 | 1 | 1 | The multiplicative reduction in the coital act rate for all relationship types when an individual has exactly three current partners. Represents coital dilution. | {
"Coital_Dilution_Factor_4_Partners": 0.45
}
|
| Condom_Transmission_Blocking_Probability | float | 0 | 1 | 0.9 | The per-act multiplier of the transmission probability when a condom is used. | {
"Condom_Transmission_Blocking_Probability": 0.99
}
|
| Heterogeneous_Infectiousness_LogNormal_Scale | float | 0 | 10 | 0 | Scale parameter of a log normal distribution that governs an infectiousness multiplier. The multiplier represents heterogeneity in infectivity and adjusts Base_Infectivity. | {
"Heterogeneous_Infectiousness_LogNormal_Scale": 1
}
|
| HIV_Adult_Survival_Shape_Parameter | float | 0.001 | 1000 | 2 | This parameter determines the shape of the Weibull distribution used to determine age-dependent survival time for individuals infected with HIV. | {
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| Immunity_Acquisition_Factor | float | 0 | 1000 | 0 | The multiplicative reduction in the probability of reacquiring disease. Only used when Enable_Immunity and Enable_Immune_Decay are set to 1. | {
"Enable_Immunity": 1,
"Enable_Immune_Decay": 1,
"Immunity_Acquisition_Factor": 0.9
}
|
| Infectivity_Exponential_Baseline | float | 0 | 1 | 0 | The scale factor applied to Base_Infectivity at the beginning of a simulation, before the infectivity begins to grow exponentially. Infectivity_Scale_Type must be set to EXPONENTIAL_FUNCTION_OF_TIME. | {
"Infectivity_Exponential_Baseline": 0.1,
"Infectivity_Exponential_Delay": 90,
"Infectivity_Exponential_Rate": 45,
"Infectivity_Scale_Type": "EXPONENTIAL_FUNCTION_OF_TIME"
}
|
| Male_To_Female_Relative_Infectivity_Multipliers | array of floats | NA | NA | 1 | An array of scale factors governing the susceptibility of females relative to males, by age. Used with Male_To_Female_Relative_Infectivity_Ages. Scaling is linearly interpolated between ages. The first value is used for individuals younger than the first age in Male_To_Female_Relative_Infectivity_Ages and the last value is used for individuals older than the last age. | {
"Male_To_Female_Relative_Infectivity_Multipliers": [
5,
1,
0.5
]
}
|
| Maternal_Transmission_ART_Multiplier | float | 0 | 1 | 0.1 | The maternal transmission multiplier for on-ART mothers. | {
"Maternal_Transmission_ART_Multiplier": 0.03
}
|
| Population_Scale_Type | enum | NA | NA | USE_INPUT_FILE | The method to use for scaling the initial population specified in the demographics input file. Possible values are:
|
{
"Population_Scale_Type": "FIXED_SCALING"
}
|
| Susceptibility_Scale_Type | enum | NA | NA | CONSTANT_SUSCEPTIBILITY | The effect of time or season on infectivity. Possible values are: CONSTANT_SUSCEPTIBILITY LOG_LINEAR_FUNCTION_OF_TIME LINEAR_FUNCTION_OF_AGE LOG_LINEAR_FUNCTION_OF_AGE |
{
"Susceptibility_Scale_Type": "CONSTANT_SUSCEPTIBILITY"
}
|
| x_Air_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by air, as provided by the migration file. Enable_Air_Migration must be set to 1. | {
"x_Air_Migration": 1
}
|
| x_Birth | float | 0 | 3.40E+38 | 1 | Scale factor for birth rate, as provided by the demographics file (see NodeAttributes parameters). Enable_Birth must be set to 1. | {
"x_Birth": 1
}
|
| x_Family_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by families, as provided by the migration file. Enable_Family_Migration must be set to true (1). | {
"x_Family_Migration": 1
}
|
| x_Local_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for rate of migration by foot travel, as provided by the migration file. Enable_Local_Migration must be set to 1. | {
"x_Local_Migration": 1
}
|
| x_Other_Mortality | float | 0 | 3.40E+38 | 1 | Scale factor for mortality from causes other than the disease being simulated, as provided by the demographics file (see Complex distributions parameters). Enable_Vital_Dynamics must be set to 1. | {
"x_Other_Mortality": 1
}
|
| x_Regional_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by road vehicle, as provided by the migration file. Enable_Regional_Migration must be set to 1. | {
"x_Regional_Migration": 1
}
|
| x_Sea_Migration | float | 0 | 3.40E+38 | 1 | Scale factor for the rate of migration by sea, as provided by the migration file. Enable_Sea_Migration must be set to 1. | {
"x_Sea_Migration": 1
}
|
Simulation setup¶
These parameters determine the basic setup of a simulation including the type of simulation you are running, such as “GENERIC_SIM” or “MALARIA_SIM”, the simulation duration, and the time step duration.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| Base_Year | float | 1900 | 2200 | 2015 | The absolute time in years when the simulation begins. This can be combined with CampaignEventByYear to trigger campaign events. | {
"Base_Year": 1960
}
|
| Config_Name | string | NA | NA | UNINITIALIZED STRING | The optional, user-supplied title naming a configuration file. | {
"Config_Name": "My_First_Config"
}
|
| Enable_Interventions | boolean | 0 | 1 | 0 | Controls whether or not campaign interventions will be used in the simulation. Set Campaign_Filename to the path of the file that contains the campaign interventions. | {
"Enable_Interventions": 1,
"Campaign_Filename": "campaign.json"
}
|
| Listed_Events | array of strings | NA | NA | [] | The list of valid, user-defined events that will be included in the campaign. Any event used in the campaign must either be one of the built-in events or in this list. | {
"Listed_Events": [
"Vaccinated",
"VaccineExpired"
]
}
|
| Memory_Usage_Halting_Threshold_Working_Set_MB | integer | 0 | 1.00E+06 | 8000 | The maximum size (in MB) of working set memory before the system throws an exception and halts. | {
"Memory_Usage_Halting_Threshold_Working_Set_MB": 4000
}
|
| Memory_Usage_Warning_Threshold_Working_Set_MB | integer | 0 | 1.00E+06 | 7000 | The maximum size (in MB) of working set memory before memory usage statistics are written to the log regardless of log level. | {
"Memory_Usage_Warning_Threshold_Working_Set_MB": 3500
}
|
| Node_Grid_Size | float | 0.00416 | 90 | 0.004167 | The spatial resolution indicating the node grid size for a simulation in degrees. | {
"Node_Grid_Size": 0.042
}
|
| Number_Basestrains | integer | 1 | 10 | 1 | The number of base strains in the simulation, such as antigenic variants. | {
"Number_Basestrains": 1
}
|
| Num_Cores | integer | NA | NA | NA | The number of cores used to run a simulation. | {
"Num_Cores": 4
}
|
| Run_Number | integer | 0 | 2147480000 | 1 | Sets the random number seed through a bit manipulation process. When running a multi-core simulation, combines with processor rank to produce independent random number streams for each process. | {
"Run_Number": 1
}
|
| Serialization_Time_Steps | array of integers | 0 | 2.15E+09 | The list of time steps after which to save the serialized state. 0 (zero) indicates the initial state before simulation, n indicates after the nth time step. By default, no serialized state is saved. | {
"Serialization_Time_Steps": [
0,
10
]
}
|
|
| Serialized_Population_Filenames | array of strings | NA | NA | NA | Array of filenames with serialized population data. The number of filenames must match the number of cores used for the simulation. The file must be in .dtk format. | {
"Serialized_Population_Filenames": [
"state-00010.dtk"
]
}
|
| Serialized_Population_Path | string | NA | NA | . | The root path for the serialized population files. | {
"Serialized_Population_Path": "../00_Generic_Version_1_save/output"
}
|
| Simulation_Duration | float | 0 | 1000000 | 1 | The elapsed time (in days) from the start to the end of a simulation. | {
"Simulation_Duration": 7300
}
|
| Simulation_Timestep | float | 0 | 1000000 | 1 | The duration of a simulation time step, in days. | {
"Simulation_Timestep": 1
}
|
| Simulation_Type | enum | NA | NA | GENERIC_SIM | The type of disease being simulated. Possible IDM-supported values are:
|
{
"Simulation_Type": "STI_SIM"
}
|
| Start_Time | float | 0 | 1000000 | 1 | The time, in days, when the simulation begins. This time is used to identify the starting values of the temporal input data, such as specifying which day’s climate values should be used for the first day of the simulation. Note The Start_Day of campaign events is in absolute time, so time relative to the beginning of the simulation depends on this parameter. |
{
"Start_Time": 365
}
|
Symptoms and diagnosis¶
The following parameters determine the characteristics of HIV diagnosis and HIV/AIDS symptoms, such as CD4 counts at various times in the progression of the disease.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| CD4_At_Death_LogLogistic_Scale | float | 1 | 1000 | 31.63 | The scale parameter of a Weibull distribution that represents the at-death CD4 cell count. | {
"CD4_At_Death_LogLogistic_Scale": 2.96
}
|
| CD4_Post_Infection_Weibull_Heterogeneity | float | 0 | 100 | 0.275642 | The inverse shape parameter of a Weibull distribution that represents the post-acute-infection CD4 cell count. | {
"CD4_Post_Infection_Weibull_Heterogeneity": 0
}
|
| CD4_Post_Infection_Weibull_Scale | float | 1 | 1000 | 560.432 | The scale parameter of a Weibull distribution that represents the post-acute-infection CD4 cell count. | {
"CD4_Post_Infection_Weibull_Scale": 550
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity | float | 0.1 | 10 | 1 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the heterogeneity (inverse shape) of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity": 0.5
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Scale | float | 1 | 3650 | 183 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the scale of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Scale": 618.3
}
|
Weibull distributions¶
The following parameters use a Weibull distribution.
| Parameter | Data type | Minimum | Maximum | Default | Description | Example |
|---|---|---|---|---|---|---|
| CD4_At_Death_LogLogistic_Heterogeneity | float | 0 | 100 | 0 | The inverse shape parameter of a Weibull distribution that represents the at-death CD4 cell count. | {
"CD4_At_Death_LogLogistic_Heterogeneity": 0.7
}
|
| CD4_At_Death_LogLogistic_Scale | float | 1 | 1000 | 31.63 | The scale parameter of a Weibull distribution that represents the at-death CD4 cell count. | {
"CD4_At_Death_LogLogistic_Scale": 2.96
}
|
| CD4_Post_Infection_Weibull_Heterogeneity | float | 0 | 100 | 0.275642 | The inverse shape parameter of a Weibull distribution that represents the post-acute-infection CD4 cell count. | {
"CD4_Post_Infection_Weibull_Heterogeneity": 0
}
|
| CD4_Post_Infection_Weibull_Scale | float | 1 | 1000 | 560.432 | The scale parameter of a Weibull distribution that represents the post-acute-infection CD4 cell count. | {
"CD4_Post_Infection_Weibull_Scale": 550
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity | float | 0.1 | 10 | 1 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the heterogeneity (inverse shape) of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Heterogeneity": 0.5
}
|
| Days_Between_Symptomatic_And_Death_Weibull_Scale | float | 1 | 3650 | 183 | The time between the onset of AIDS symptoms and death is sampled from a Weibull distribution; this parameter governs the scale of the Weibull. | {
"Days_Between_Symptomatic_And_Death_Weibull_Scale": 618.3
}
|
| HIV_Adult_Survival_Scale_Parameter_Intercept | float | 0.001 | 1000 | 21.182 | This parameter determines the intercept of the scale parameter, λ, for the Weibull distribution used to determine HIV survival time. Survival time with untreated HIV infection depends on the age of the individual at the time of infection, and is drawn from a Weibull distribution with shape parameter (see HIV_Adult_Survival_Shape_Parameter) and scale parameter, λ. The scale parameter is allowed to vary linearly with age as follows: λ = HIV_Adult_Survival_Scale_Parameter_Intercept + HIV_Adult_Survival_Scale_ Parameter_Slope * Age (in years). |
{
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Adult_Survival_Scale_Parameter_Slope | float | -1000 | 1000 | -0.2717 | This parameter determines the slope of the scale parameter, λ, for the Weibull distribution used to determine HIV survival time. Survival time with untreated HIV infection depends on the age of the individual at the time of infection, and is drawn from a Weibull distribution with shape parameter (see HIV_Adult_Survival_Shape_Parameter) and scale parameter, λ. The scale parameter is allowed to vary linearly with age as follows: λ = HIV_Adult_Survival_Scale_Parameter_Intercept + HIV_Adult_Survival_Scale_ Parameter_Slope * Age (in years). Because survival time with HIV becomes shorter with increasing age, HIV_Adult_Survival_Scale_ Parameter_Slope should be set to a negative number. |
{
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Adult_Survival_Shape_Parameter | float | 0.001 | 1000 | 2 | This parameter determines the shape of the Weibull distribution used to determine age-dependent survival time for individuals infected with HIV. | {
"HIV_Adult_Survival_Scale_Parameter_Intercept": 21.182,
"HIV_Adult_Survival_Scale_Parameter_Slope": -0.2717,
"HIV_Adult_Survival_Shape_Parameter": 2.0,
"HIV_Age_Max_for_Adult_Age_Dependent_Survival": 50.0
}
|
| HIV_Child_Survival_Slow_Progressor_Scale | float | 0.001 | 1000 | 16 | The Weibull scale parameter describing the distribution of HIV survival for children who are slower progressors. | {
"HIV_Child_Survival_Slow_Progressor_Scale": 16.0,
"HIV_Child_Survival_Slow_Progressor_Shape": 2.7
}
|
| HIV_Child_Survival_Slow_Progressor_Shape | float | 0.001 | 1000 | 2.7 | The Weibull shape parameter describing the distribution of HIV survival for children who are slower progressors. | {
"HIV_Child_Survival_Slow_Progressor_Scale": 16.0,
"HIV_Child_Survival_Slow_Progressor_Shape": 2.7
}
|
| Sexual_Debut_Age_Female_Weibull_Heterogeneity | float | 0 | 50 | 20 | The inverse shape of the Weibull distribution for female debut age. | {
"Sexual_Debut_Age_Female_Weibull_Heterogeneity": 0.05
}
|
| Sexual_Debut_Age_Female_Weibull_Scale | float | 0 | 50 | 16 | The scale term of the Weibull distribution for female debut age. | {
"Sexual_Debut_Age_Female_Weibull_Scale": 15.919654846191
}
|
| Sexual_Debut_Age_Male_Weibull_Heterogeneity | float | 0 | 50 | 20 | The inverse shape of the Weibull distribution for male debut age. | {
"Sexual_Debut_Age_Male_Weibull_Heterogeneity": 0.05
}
|
| Sexual_Debut_Age_Male_Weibull_Scale | float | 0 | 50 | 16 | The scale term of the Weibull distribution for male debut age. | {
"Sexual_Debut_Age_Male_Weibull_Scale": 16.946729660034
}
|
| Sexual_Debut_Age_Min | float | 0 | 3.40E+38 | 13 | The minimum age at which individuals become eligible to form sexual relationships. | {
"Sexual_Debut_Age_Min": 13
}
|
 matrix must be specified in the demographics file (see
matrix must be specified in the demographics file (see