Malaria model overview¶
The Epidemiological MODeling software (EMOD), malaria model, is an agent-based, discrete time, Monte Carlo simulator used to simulate malaria conditions in order to evaluate the effectiveness of eradication or mitigation approaches. The model utilizes microsolvers to combine detailed vector population dynamics, human population dynamics, human immunity, within-host parasite dynamics, effects of antimalarial drugs, and other aspects of malaria biology to simulate malaria transmission. EMOD can be calibrated to particular geographic locations, and the microsolver framework enables the model’s functionality to be highly modifiable. Further, the framework includes the ability to add intervention campaigns, and those interventions can be specified to target particular populations or sub-populations of human or vector groups. Interventions can be deployed within simulations for a variety of transmission settings with different transmission intensities, vector behaviors, and seasonally-driven ecologies.
The following pages describe the structure of the model, and then explore each of the model components. Additionally, the specifics of the model are discussed in detail in the articles Eckhoff, Malaria Journal 2011, 10:303; Eckhoff, Malaria Journal 2012, 11:419; Eckhoff, PLoS ONE 2012, 7(9); and Eckhoff, Am. J. Trop. Med. Hyg. 2013, 88(5).
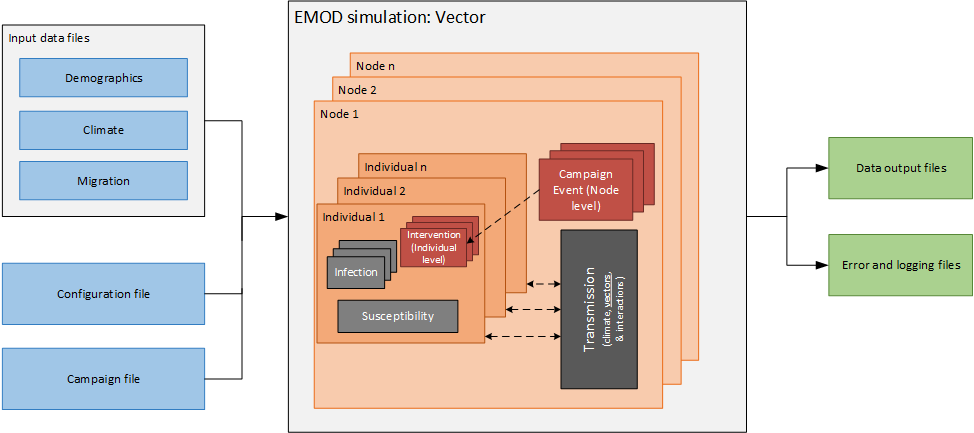
The generic software architecture of EMOD can be found on Introduction to the software. The malaria EMOD is based on the same architecture, with modifications that transform the generic model into a vector-based model. For example, transmission occurs via vectors (and the associated climatalogical impacts on vector biology), individuals can have multiple infections, and campaign interventions can act at the individual level or at the node level.
Typical SEIR models are described in Introduction to disease modeling. Vectors add a level of complexity to the interactions, as pathogens are transmitted via vector - human - vector. For malaria, the SEIR model would appear as follows:
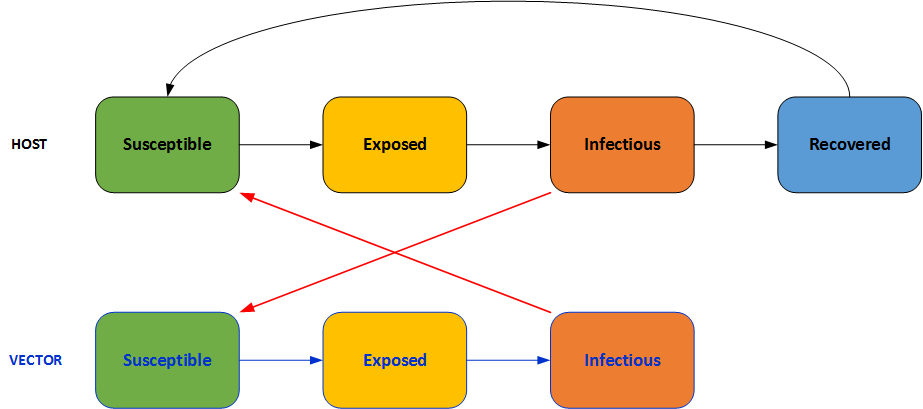
While EMOD is an agent-based model, both the simulated humans and vectors move through the various infection states analogously to the compartmental model illustrated above. To account for the real-world complexity of malaria transmission, numerous parameters have been added to EMOD to increase its predictive power. Further, these parameters and their associated microsolvers allow EMOD to model aspects of malaria infections and population dynamics that do not readily fit into the SEIRS framework (for example, within-host parasite dynamics, parasitalogical immunity, and innate and adaptive responses to antigens). Finally, as an agent- based model, EMOD enables the addition of spatial and temporal properties to the simulation. The following pages describe this complexity in detail.