Generic 6: SEIRS - Disease-specific mortality and endemics¶
This tutorial assumes that you have read Generic 5: SIRS - Waning immunity.
This tutorial is a simple SEIRS model in a single-node, village-scale location. Similar to the SIRS model, recovered individuals transition to susceptible individuals by enabling waning immunity.
As in a previous tutorial, Generic 4: SEIR - Vital dynamics (birth and death), the vital dynamics are enabled. This scenario also enables the disease-specific mortality rate.
By varying the disease infectivity and infection durations, you will observe the effect of these parameters on the height of the epidemic peak, the time to reach the peak, and the cumulative number of disease cases.
Demographics inputs¶
This scenario uses generic_scenarios_demographics file for demographics input. You can view the complete demographics file at <path_to_directory>RegressionScenariosInputFiles where <path_to_directory> is the location where EMOD source files were installed. For more information on demographics files, see Demographics parameters.
Key configuration parameters¶
You can view the complete config.json in the <path_to_directory>RegressionScenariosGeneric06_SEIRS directory.
Population dynamics parameters¶
The following parameters enable vital dynamics for birth , non-disease related deaths, and disease related deaths. For more information, see Population dynamics parameters.
Set the following parameters:
- Set Enable_Vital_Dynamics to 1.
- Set Enable_Birth to 1.
- Set Death_Rate_Dependence to “NONDISEASE_MORTALITY_BY_AGE_AND_GENDER.”
- Set Enable_Disease_Mortality to 1.
SEIRS model setup parameters¶
The incubation period, infection period, immunity, and immunity waning must be enabled for an SEIRS model. For more information, see Incubation and Immunity parameters.
Set the following parameters:
- Set Base_Incubation_Period to 3.
- Set Enable_Immunity to 1.
- Set Enable_Immune_Decay to 1.
- Set Incubation_Period_Distribution to “EXPONENTIAL_DISTRIBUTION.”
Disease parameters¶
These parameters set the EMOD executable to run as an SEIRS-like disease model with person-to- person direct contact. For more information, see Infectivity and transmission parameters.
Set the following parameters:
- Set Base_Infectivity to 0.25.
- Set Base_Infectious_Period to 6.
- Set Infectious_Period_Distribution to “EXPONENTIAL_DURATION.”
Mortality rate¶
The following parameters set the mortality for the disease. These parameters determine whether disease mortality is enabled by the simulation when the mortality is applied, and the rate of mortality. For more information, see Mortality and survival parameters.
Set the following parameters:
- Set Base_Mortality to 0.01.
- Set Mortality_Time_Course to “DAILY_MORTALITY.”
Immunity parameters¶
For this scenario the Mortality_Blocking_Immunity_Duration_Before_Decay and Mortality_Blocking_Immunity_Decay_Rate are added. On recovery, people’s immunity decays with an average half-life of 10 days and a 100-day offset. This applies to transmission blocking, acquisition blocking and mortality blocking immunity. For more information, see Immunity parameters.
Set the following parameters:
- Set Acquisition_Blocking_Immunity_Duration_Before_Decay to 100.
- Set Acquisition_Blocking_Immunity_Decay_Rate to 0.1.
- Set Transmission_Blocking_Immunity_Duration_Before_Decay to 100.
- Set Transmission_Blocking_Immunity_Decay_Rate to 0.1.
- Set Immunity_Acquisition_Factor to 0.
- Set Immunity_Mortality_Factor to 0.
- Set Immunity_Transmission_Factor to 0.
- Set Mortality_Blocking_Immunity_Duration_Before_Decay to 100.
- Set Mortality_Blocking_Immunity_Decay_Rate to 0.1.
Simulation duration parameters¶
The simulation is run for 2 years with a 1-day time step. For more information, see Simulation setup parameters.
Set the following parameters:
- Set Simulation_Timestep to 1.
- Set Simulation_Duration to 730.
Interventions¶
You can view the complete campaign.json in the SamplesGeneric06_SEIRS directory.
Baseline outbreak¶
This scenario uses an OutbreakIndividual event at day 1 as the initial seeding of the infection. The value of Demographic_Coverage is 0.001 which is about 10 infected individuals in a population of 10,000. For more information, see OutbreakIndividual parameters.
{
"Event_Coordinator_Config": {
"Intervention_Config": {
"Antigen": 0,
"Genome": 0,
"Outbreak_Source": "PrevalenceIncrease",
"class": "OutbreakIndividual"
},
"Target_Demographic": "Everyone",
"Demographic_Coverage": 0.001,
"class": "StandardInterventionDistributionEventCoordinator"
},
"Event_Name": "Outbreak",
"Nodeset_Config": {
"class": "NodeSetAll"
},
"Start_Day": 1,
"class": "CampaignEvent"
}
Vaccination campaigns¶
The following campaign event adds a vaccination campaign at day 500, with 100% vaccine take rate (Vaccine_Take) and 80% coverage (Demographic_Coverage) for all individuals. The vaccine is acquisition-blocking and full immunity lasts for 10 years (3650 days). For more information, see SimpleVaccine parameters.
{
"Use_Defaults": 1,
"Events": [{
"Event_Coordinator_Config": {
"Demographic_Coverage": 0.80,
"Intervention_Config": {
"Cost_To_Consumer": 10.0,
"Durability_Time_Profile": "BOXDURABILITY",
"Primary_Decay_Time_Constant": 3650,
"Reduced_Acquire": 1,
"Reduced_Transmit": 0,
"Vaccine_Take": 1,
"Vaccine_Type": "AcquisitionBlocking",
"class": "SimpleVaccine"
},
"Target_Demographic": "Everyone",
"class": "StandardInterventionDistributionEventCoordinator"
},
"Nodeset_Config": {
"class": "NodeSetAll"
},
"Start_Day": 5060,
"class": "CampaignEvent"
}]
}
Run the simulation¶
Run the simulation and generate graphs of the simulation output. For more information, see Run simulations.
Note
Because the EMOD model is stochastic, your graphs may appear slightly different from those given below.
Simulation output graphs¶
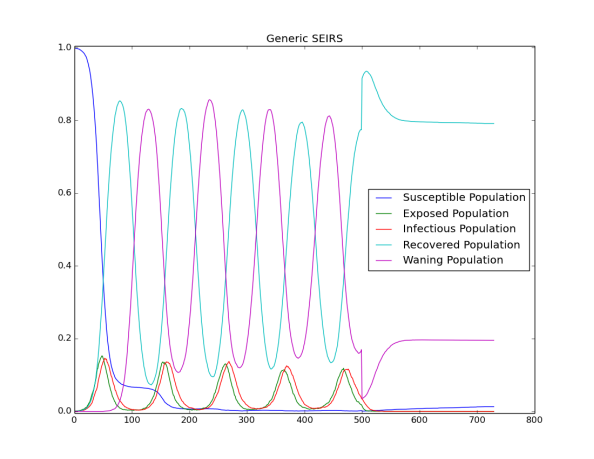
The following figure shows the complete trajectory of the outbreak: the disease endemicity due to vital process and waning immunity; and the effect of vaccination campaigns that eradicates the outbreak after day 500.
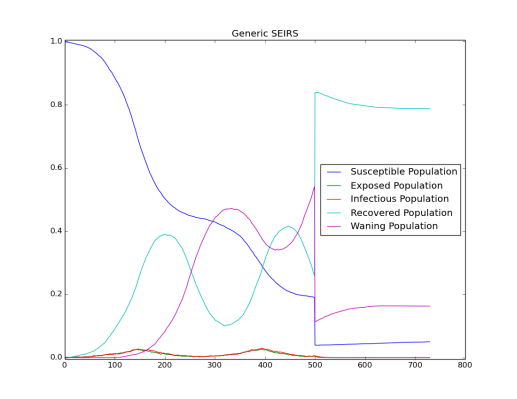
Figure 1: Trajectory of the outbreak
The following figure shows the number of disease-specific deaths, and cumulative number of infections for all of the channels in the simulation output.
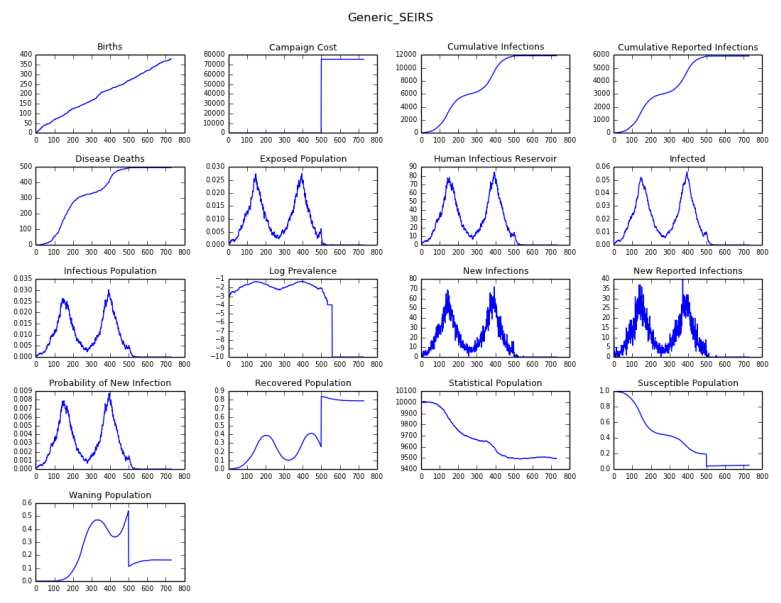
Figure 2: Disease-specific deaths and cumulative number of infections
Exploring the model¶
In the this simulation, the Base_Infectivity parameter is set to 0.25 and with the incubation period, the basic reproductive numbers is 1.5. Change the Base_Infectivity parameter to 0.5. The base reproductive number should now be 3. Due to this larger value, the disease did not persist in the simulation because the rate of consuming susceptible individuals was larger than the inflow of new susceptible individuals. The infection could not find enough new susceptible individuals to sustain the transmission.
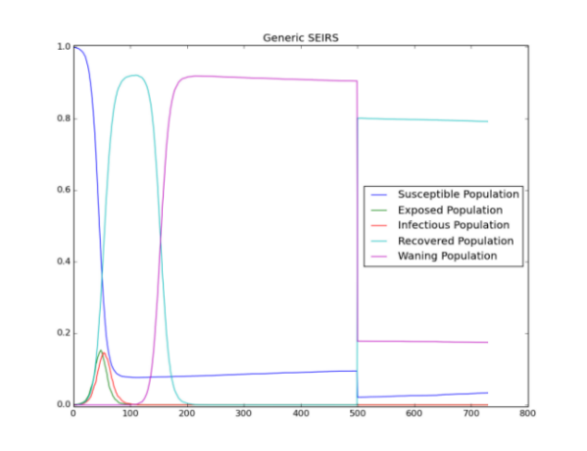
Keep the base reproductive number at 3, and try shortening the immunity duration by changing Acquisition_Blocking_Immunity_Duration_Before_Decay and Transmission_Blocking_Immunity_Duration_Before_Decay from 100 to 50. Run the simulation again, and observe the persistence of the outbreak again due to increased influx of susceptible individuals.